-
桂花Osmanthus fragrans是中国十大传统名花之一,树形优美,花香怡人,兼具观赏、食用、药用、香用等价值,被广泛应用于园林绿化以及食品和护肤品等加工产业[1−2]。然而,由于桂花花期较短,最佳观赏期和采收期仅2~3 d,且花朵小,易褐化,制约了桂花价值的开发利用[3]。因此,研究桂花花瓣衰老及调控机制,对延长花期、提升品质、促进产业应用具有重要意义。
有研究表明:在花瓣衰老层面,许多经济采收价值较高的不结实桂花品种对乙烯敏感,乙烯参与了衰老过程中花瓣脱落和萎蔫、细胞结构变化、氧化还原系统及核酸降解等多个过程的调节,是桂花衰老的重要调控因子[4−7]。桂花开花多组学联合分析发现:小生长素上调RNA (small auxin up-regulated RNA, SAURs)和乙烯响应因子 (ethylene-responsive ranscription factors, ERFs)分别在开花和衰老阶段富集,可能参与调控衰老进程[8]。ERFs转录因子广泛参与植物衰老调控的机制也较为成熟[9]。但在桂花中缺乏SAUR基因的相关报道。
SAUR基因家族是植物特有的,生长素早期响应基因中最大的1个家族,SAUR基因广泛存在于各种植物中,如拟南芥Arabidopsis thaliana有79个,水稻Oryza sativa有58个(包括2个假基因),高粱Orghum bicolor有71个,马铃薯Solanum tuberosum有134个,番茄Solanum lycopersicum有99个,玉米Zea mays有91个,棉花Gossypium hirsutum有632个,苹果Malus domestica有80个,甜樱桃Prunus avium有86个等[10]。SAUR被报道参与调节多种生物过程[11]。研究发现:SAUR与生长素介导的细胞扩增密切相关,广泛参与生长素的合成和运输[12],改变顶沟和子叶发育[13],促进叶片生长和衰老[14]、果实成熟或根生长和发育[15]。在拟南芥中,AtSAUR36和AtSAUR41的表达水平与细胞扩张密切相关,过量表达会显著促进下胚轴表皮细胞的伸长[16],而且SAUR32、SAUR19和SAUR36主要与顶端弯钩的形成相关,过量表达会使幼苗下胚轴较短[17]。AtSAUR41、AtSAUR76与根的发育相关,它们的转录上调能促进主根伸长和侧根发育[18]。杨树Populus tomentosa中SAUR12、SAUR34、SAUR54、SAUR67、SAUR91和SAUR97的上调表达提高了幼苗对低温的适应性[19]。此外,SAUR基因家族蛋白受miR159调控,会增强小麦Triticum aestivum的耐旱性[20]。而且,SAUR30也与杨树的干旱适应相关[21]。在番茄中,SlSAUR69基因的过表达增强了番茄对乙烯的敏感性,从而促进了果实成熟[22]。除生长素外,乙烯、油菜素内酯、赤霉素、脱落酸、茉莉酸、光和渗透胁迫,以及一些关键开花相关转录因子也调控SAUR基因的表达[23],表明SAUR参与了激素和环境因子介导的植物生长发育及开花调控。
前期对桂花开花衰老相关基因的多组学联合分析研究中,已筛选获得21个SAUR家族基因,其中OfSAUR21 (基因登录号:LYG019593)是仅在花器官中特异表达的基因成员[8]。本研究在此基础上,首先对OfSAUR21基因进行克隆,随后运用生物信息学分析基因分子特性,构建该基因的超表达载体,同时通过瞬时转化桂花花瓣的方式开展基因功能验证,以期为解析桂花开花衰老的调控机制提供理论参考。
-
所用材料‘柳叶金桂’O. fragrans ‘Liuye Jingui’采自华中农业大学校园(30°29′N,114°21′E)。对桂花的6个开花时期(铃梗期、初花期、盛花初期、盛花期、盛花末期、衰老期)进行采样(图1)。对采集样品称量后,密封保存于液氮中,立即运回实验室保存在−80℃冰箱内,以备后续分析。瞬时转化以四季桂O. fragrans ‘Sijigui’为材料[24]。
-
根据桂花数据库获取OfSAUR21的编码序列(CDS),设计全长引物(由武汉擎科生物有限公司合成,表1)。分别提取桂花不同开花时期的样品,液氮研磨后,按植物RNA小量提取试剂盒说明书提取RNA,再按cDNA逆转录试剂盒说明书获得cDNA后加无酶水稀释10或20倍。取 4 μL 6个开花阶段混样cDNA模板配制 40 μL PCR体系:2×Trans Taq-T PCR Super Mix 20 μL,目的基因上下游引物各0.5 μL,双氧水(ddH2O) 15 μL。PCR反应程序:94 ℃预变性5 min,94 ℃变性30 s,68 ℃退火30 s,72 ℃延伸2 min,共35个循环。用琼脂糖凝胶电泳进行检测,确定条带后,用琼脂糖凝胶回收试剂盒回收目的DNA。然后连接到pTOPO-T载体,转化大肠埃希菌Escherichia coli DH5α感受态细胞,筛选阳性克隆,在武汉擎科生物有限公司进行基因测序。
引物名称 引物序列 (5′→3′) OfSAUR21-F GGTACCATGGCCATCCGCATGCCTCGTAT OfSAUR21-R GTCGACCTATATTACACCCAACTGTGAAATGATA OfRAN1-F AGAACCGACAGGTGAAGGCAA OfRAN1-R TGGCAAGGTACAGAAAGGGCT OfSAUR21-Q-F TGACGGGAAACCAAACAATGATAGAT OfSAUR21-Q-R TAATAAATGTATCCTCGCAACAGGGGA Table 1. Primers used in the study
-
用美国国家生物技术信息中心(NCBI)、DNA分析软件(DNAMAN)、分子进化遗传分析软件(MEGA7.0)等,查找OfSAUR21同源基因序列,构建进化树,进行氨基酸比对;通过ORFfinder系统(https://www.ncbi.nlm.nih.gov/orffinder)预测基因开放阅读框与保守结构域;借助lncLocator在线工具(http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/)分析基因亚细胞定位;利用ExPASy ProtParam工具(https://web.expasy.org/protparam/)解析该基因编码蛋白质的理化性质;通过SOPMA在线程序(http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html)预测蛋白质二级结构;并通过SWISS-MODEL交互式平台(https://swissmodel.expasy.org/interactive)构建OfSAUR21蛋白的三级结构模型。用ProtScale预测OfSAUR21氨基酸序列的疏水性与亲水性。
-
根据获得的桂花OfSAUR21基因的序列非保守区域设计定量引物(表1),利用实时荧光定量PCR (RT-qPCR) 技术分析不同开花时期的表达模式。参考之前的研究[25],RT-qPCR反应在Line Gene 9600荧光定量PCR检测系统上应用SYBR®Premix Ex TaqTM进行。PCR反应的总体积为20 µL,包括10 µL SYBR Green PCR混合物、2 µL cDNA、0.8 µL正向和反向引物、6.4 µL ddH2O。PCR反应遵循以下程序:94 ℃ 30 s 1个循环,94 ℃ 10 s和60 ℃ 30 s,40个循环,桂花RAN1基因用作内参。将铃梗期样品的表达水平设定为1,用2−ΔΔCt方法标准化相对表达水平[26]。
-
测序后,将序列比对正确的菌液进行复苏,提取目的基因和空载体(P2300)质粒。配制50 μL双酶切体系:10×Green Buffer 5 μL,SalⅠ 1 μL,KpnⅠ 1 μL,质粒 25 μL,ddH2O 18 μL,将反应体系放入水浴锅中,37 ℃反应3 h后进行切胶回收。配制10 μL T4连接酶体系:1 μL 10×Buffer,1 μL T4 DNA连接酶,2 μL P2300质粒,6 μL目的基因质粒,在电子水浴锅22 ℃恒温连接1~2 h。取50 μL大肠埃希菌DH5α,向其中加入5 μL连接产物,提取质粒。将1 μL提取的质粒加入到25 μL农杆菌 Agrobacterium tumefaciens GV3101中,经PCR验证正确后用于转化实验。
-
采集桂花初花期的花进行转基因实验。将OfSAUR21全长克隆到pCAMBIA2300s载体中,并以空的pCAMBIA2300s载体作为阴性对照。转基因条件参照文献[27],接种后低温暗培3 d,立刻收集花瓣进行RT-qPCR和乙烯测定。RT-qPCR方法参见1.2.3。乙烯测定参照文献[16],用气相色谱分析仪测定乙烯释放量,测定的乙烯标样浓度(x)与出峰面积(y)的标准曲线方程为y=81.539x−20.003。
-
采用SPSS软件进行统计分析,通过单因素方差分析(one-way ANOVA)检验组间差异显著性,随后进行Duncan多重比较,显著性水平为0.05。本研究的桂花为自然花期6个开花阶段的基因组和转录组测序数据,参考了ZOU等[28]的研究。每个处理设3个重复。所有数据均以平均值±标准差表示。
-
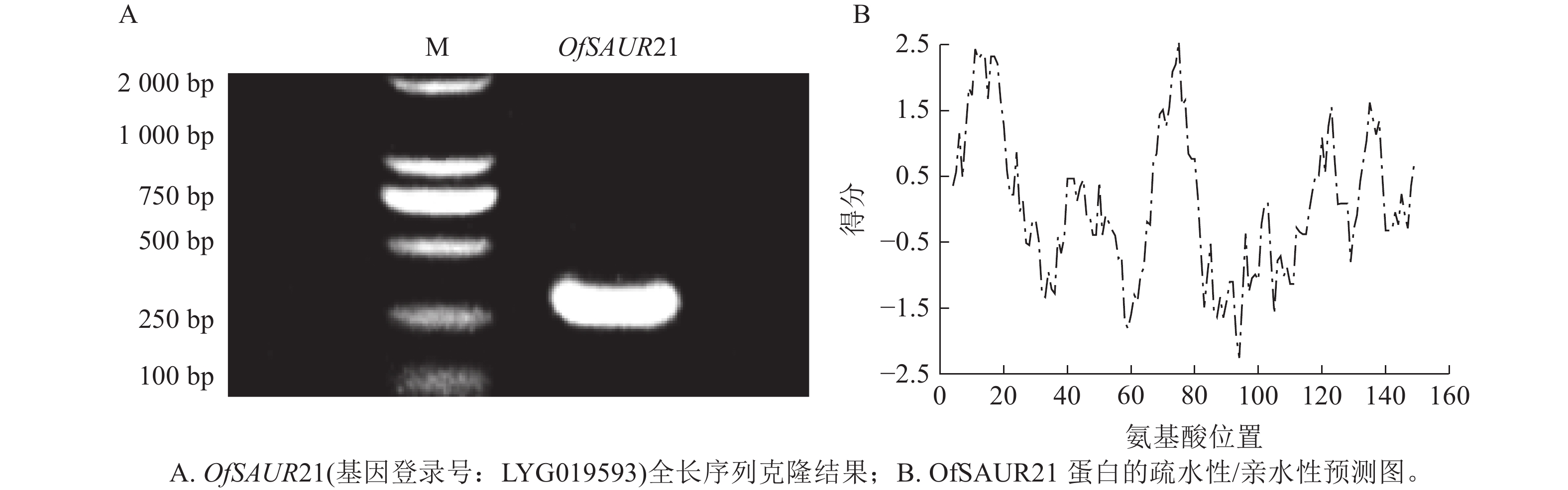
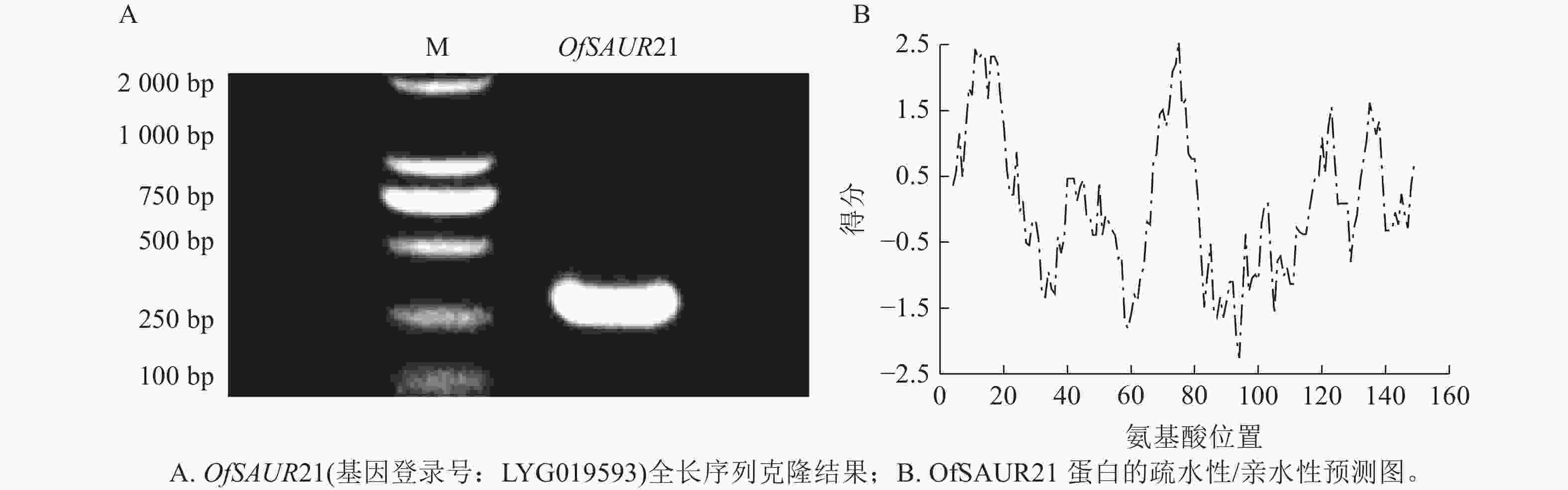
以桂花花瓣cDNA为模板,PCR扩增获得长度为294 bp的OfSAUR21基因cDNA序列(图2A)。ProtParam分析显示:OfSAUR21基因编码的蛋白质含97个氨基酸,分子式为C499H775N137O138S6,分子量为11 093.84 Da,等电点为6.57,正/负电荷残基数为8/9,脂肪系数为92.37,总平均亲水性为−0.063,不稳定性系数为43.73。ProtScale预测显示:OfSAUR21蛋白疏水峰大于0的氨基酸占比不足50%,整体呈亲水性(图2B)。在线软件https://wolfpsort.hgc.jp/预测其亚细胞定位在叶绿体。二级结构预测表明:α-螺旋占比37.11%,β-转角占比6.19%,无规则卷曲占比40.21%,延伸链占比16.49%。NCBI预测OfSAUR21结构域属于生长素响应基因家族。
-
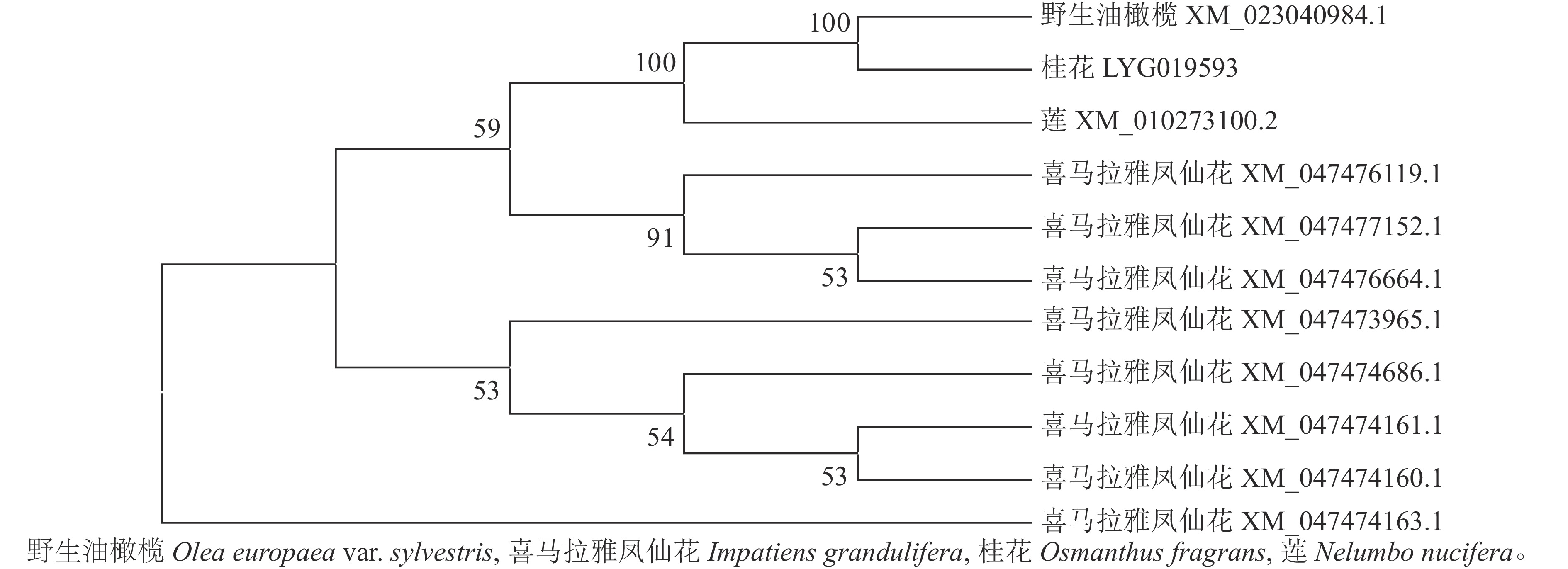
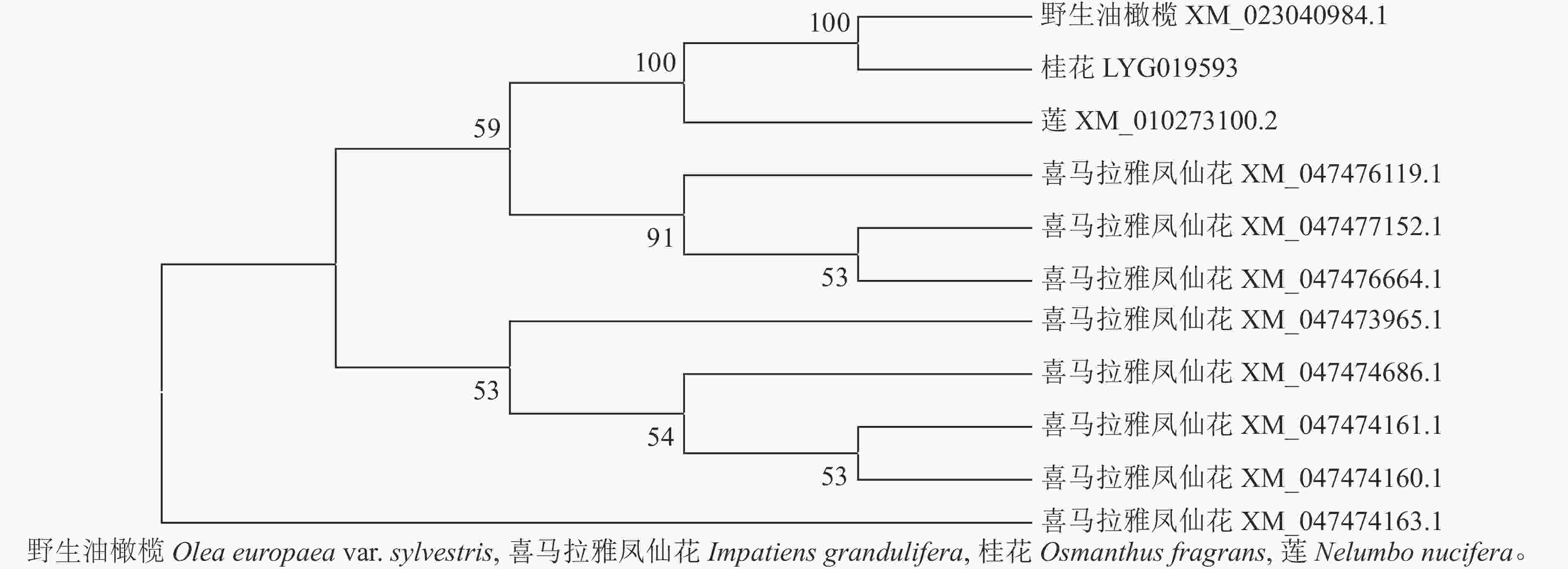
首先用NCBI上的BLAST进行序列同源性比对分析。从图3可以看出:该序列与野生油橄榄Olea europaea var. sylvestris XM_023040984.1、喜马拉雅凤仙花Impatiens glandulifera XM_047477152.1等8个同源序列及莲Nelumbo nucifera XM_010273100.2的同源性为75.88%~80.82%,其中与野生油橄榄树 XM_023040984.1的同源性最高。利用MEGA 7.0软件对该基因与上述高同源性物种(野生油橄榄、喜马拉雅凤仙花、莲)的蛋白序列进行系统进化树分析(图4)显示:OfSAUR21蛋白与野生油橄榄的亲缘关系最近。
-
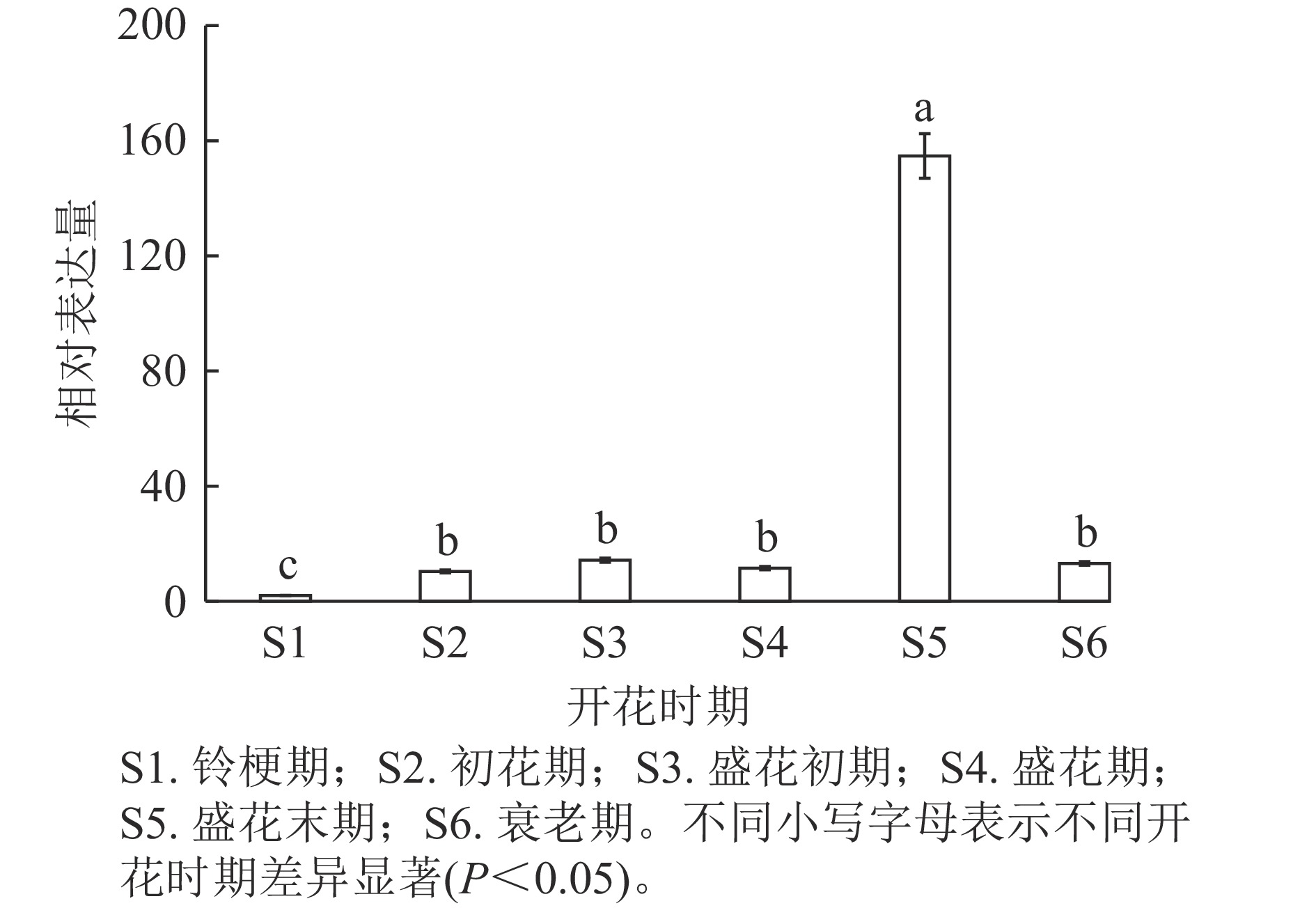
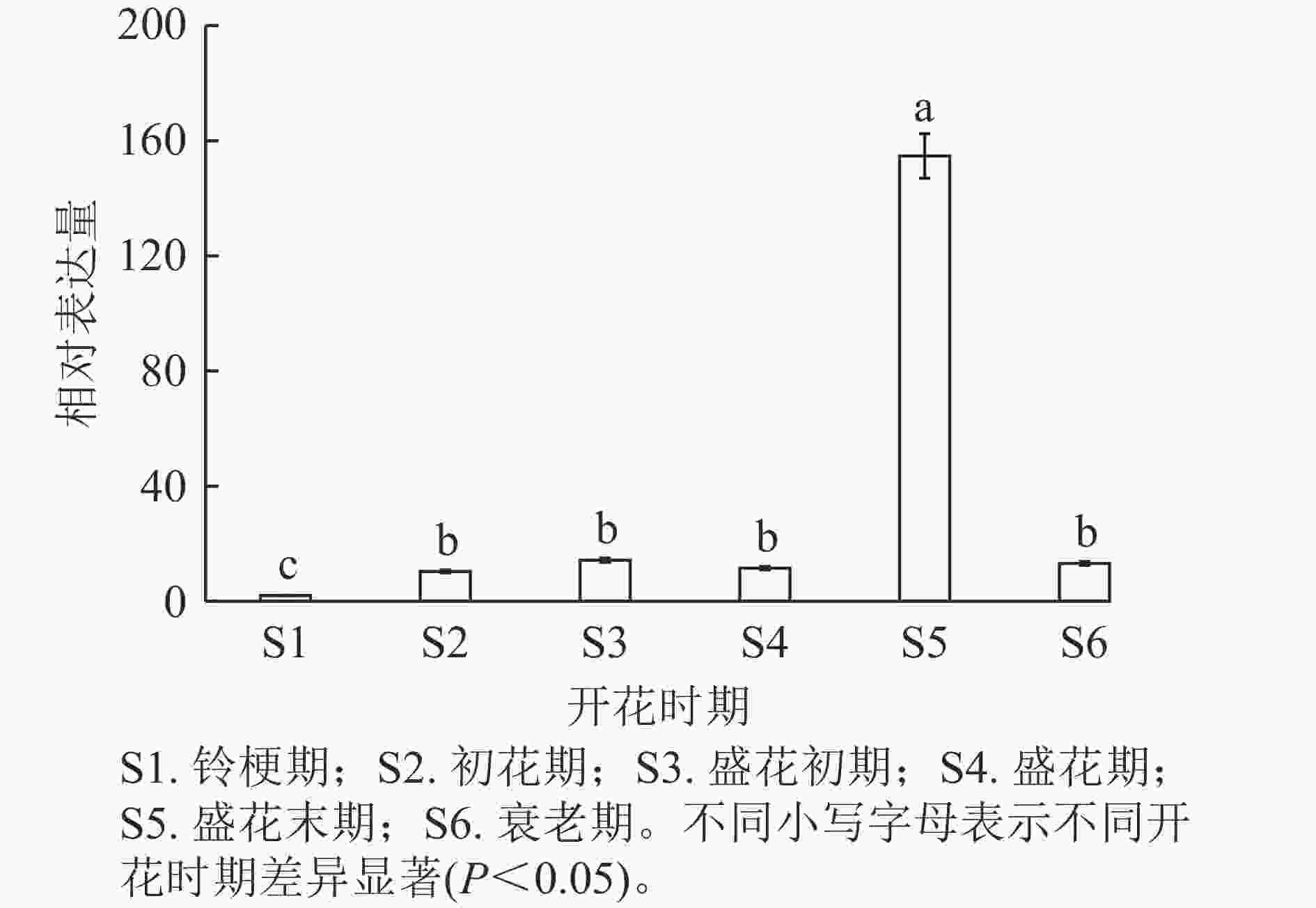
通过分析OfSAUR21基因在不同开花时期的表达模式发现:在盛花末期,花瓣中OfSAUR21基因表达量最高,显著高于其他时期(P<0.05)。而在其他5个时期中,除了铃梗期表达量最低外,其他时期花瓣中OfSAUR21基因表达量差异不显著(图5)。由此可知:OfSAUR21基因在开花中后期的表达量明显增加,表明其很可能参与了花瓣的衰老过程。
-
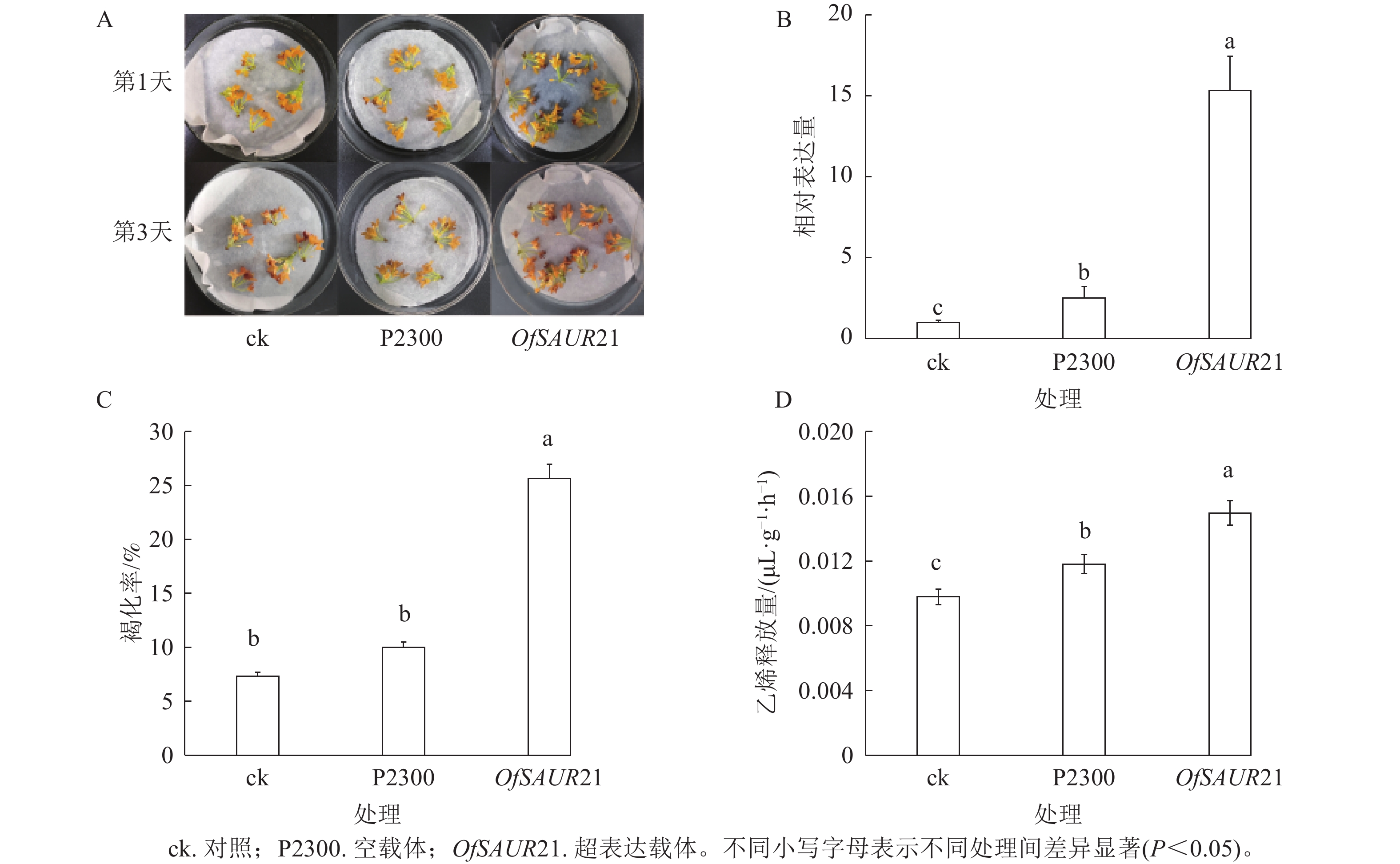
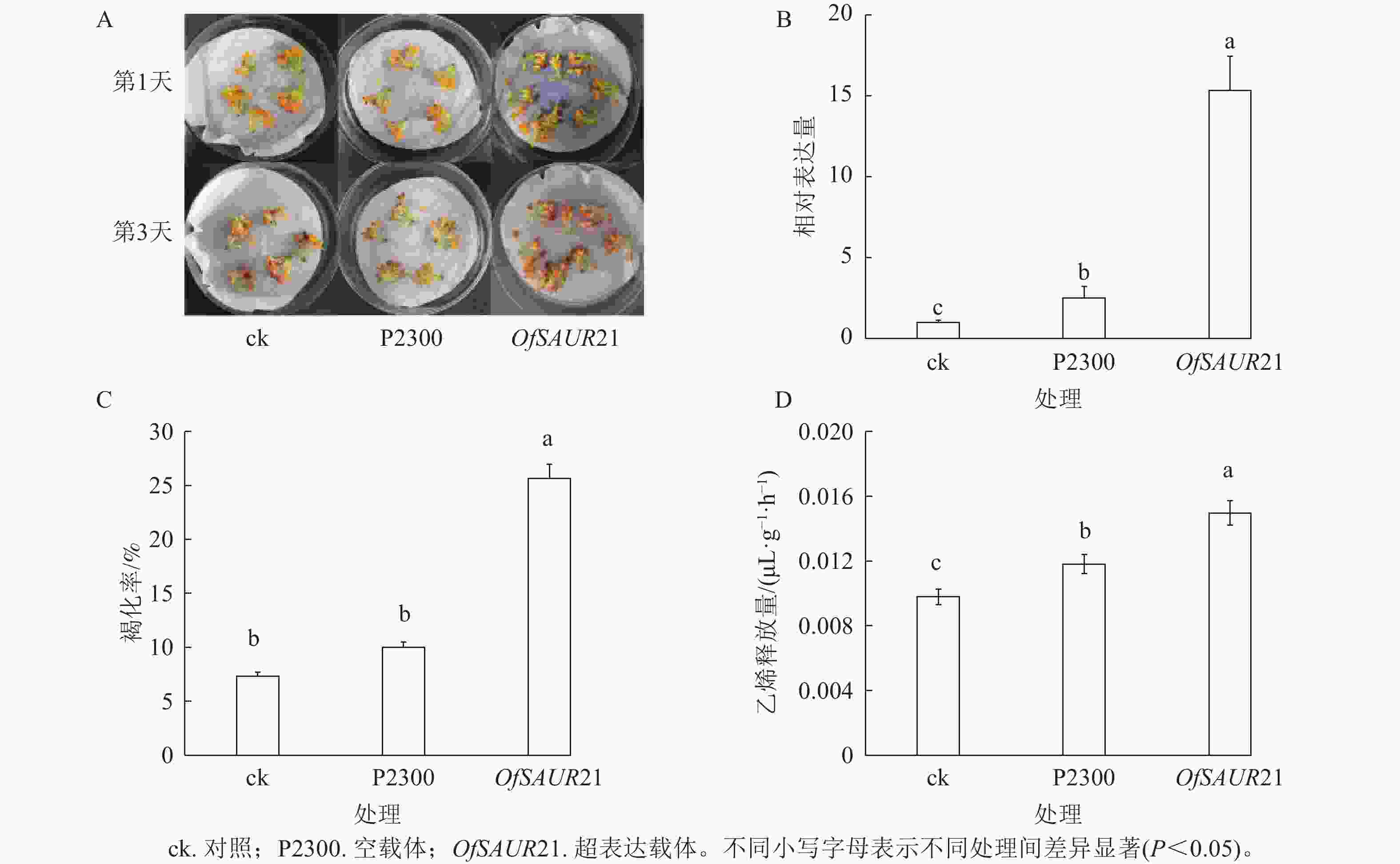
为探究OfSAUR21基因对桂花衰老的影响,开展了瞬时转化实验。如图6A所示:将OfSAUR21基因超表达载体、空载体分别导入桂花花瓣,并以未处理花瓣作为对照组,结果显示:与空载体、对照相比,超表达载体的褐化现象更为严重。3组样品RT-qPCR分析发现:不同样本在桂花花瓣中的表达量存在显著差异(P<0.05)。图6B表明:OfSAUR21基因的表达量均高于空载体、对照。对3组样品的褐化率和乙烯释放量比较发现:OfSAUR21超表达组与空载体、对照之间的褐化率(图6C)和乙烯释放量存在显著差异(图6D)(P<0.05)。综上,OfSAUR21基因的超表达提高了桂花花瓣的褐化率和乙烯释放量,促进了桂花的衰老进程。
-
SAUR基因家族作为植物特有的家族,不仅是生长素早期响应基因中规模最大的家族,更在植物生长发育的复杂调控网络中占据关键地位,广泛参与植物的多种生命活动过程,包括器官发生、组织分化、逆境适应等[10−15]。大量研究证实:SAUR基因家族的核心作用是通过精准调控细胞的膨大与扩展过程,深刻影响植物的生长发育[15]。在模式植物拟南芥中,SPARTZ等[29]研究发现:AtSAUR19~AtSAUR24基因亚组能够通过显著促进细胞的膨大生长,直接调控幼苗下胚轴的伸长和叶片的扩展发育,揭示了SAURs在植物营养生长阶段的重要调控作用。此外,拟南芥中其他SAURs成员的功能研究也呈现出丰富的生物学效应,例如AtSAUR10、AtSAUR36、AtSAUR72等基因的过表达会引发植物出现明显的过早衰老表型,包括叶片黄化加速、植株生命周期缩短等[30−31],暗示SAUR基因家族在植物衰老进程中可能扮演着复杂的调控角色。在单子叶模式植物水稻中,相关研究同样印证了SAURs基因与衰老调控的密切关联。有研究发现:OsSAUR39基因的过表达会导致水稻叶片叶绿素含量显著下降,同时加速叶片的衰老进程,表明SAUR基因家族成员在不同植物类群中可能保守地参与衰老调控[32]。综合不同物种中SAUR基因家族成员的功能研究结果,可以看出该家族基因在植物器官衰老调控过程中发挥的重要作用具有一定的普遍性,这为探索植物衰老的分子机制提供了重要方向。
本研究显示:OfSAUR21包含生长素响应家族蛋白的典型保守结构域,这与SAUR基因家族成员的核心特征高度一致,提示其可能通过响应生长素信号参与细胞生理活动的调控。OfSAUR21基因与油橄榄SAUR21的高同源性,不仅暗示了在进化历程中,这一基因在木犀科Oleaceae植物中可能具有较近的共同祖先,更提示其功能或许存在一定的保守性。油橄榄与桂花同属木犀科,在生长习性、器官发育模式上存在诸多相似之处,这种同源性为后续研究OfSAUR21在木犀科植物中的共性功能提供了重要的生物学基础。基因表达的时空特异性是功能行使的重要特征。本研究发现:OfSAUR21在桂花开花后期的表达量显著上调,这一结果与花瓣衰老进程呈现出高度的时间关联性有关。开花后期是桂花花瓣从完全绽放走向衰败的关键转折期,此时花瓣细胞的生理代谢发生剧烈变化,包括细胞膜透性增加、抗氧化系统失衡等[4]。OfSAUR21在这一时期的强势表达,很可能是它参与调控这些生理过程的直接体现。
为验证这一推测,进一步开展了桂花花瓣瞬时转化功能分析实验。本研究结果表明:与对照相比,成功侵染并过表达OfSAUR21基因的桂花花瓣,褐化率明显提高,同时花瓣组织中的乙烯释放量也显著增加。这一结果为OfSAUR21参与桂花花瓣衰老调控提供了直接的功能证据。朱诚等[33]研究指出:植物花寿命的调控可能通过调节乙烯在植物体内的产生速率,以及维持活性氧的产生与清除之间的动态平衡来实现的。而陈洪国等[34]研究发现:在桂花开花和衰老的整个进程中,乙烯释放量呈现明显的规律性变化,因此乙烯释放量可作为判断桂花开花进程与衰老程度的重要生理指标。结合已有研究证实:乙烯的产生是桂花开花和衰老的主要生理原因[35],本研究推测:OfSAUR21基因可能通过促进乙烯的释放,成为调控桂花花瓣衰老的正向调节因子。目前在拟南芥等模式植物中,尚未发现SAUR21基因直接参与衰老调控的实验证据。此外,已有研究表明:SAUR21基因可能参与植物激素信号传导过程[36],这为解释本研究中过表达OfSAUR21能促进乙烯生成提供了潜在的分子机制线索,暗示OfSAUR21基因可能通过整合生长素与乙烯等多种激素信号,实现对花瓣衰老的精细调控。当然,关于OfSAUR21基因的功能解析仍需深入。未来研究将通过建立稳定的遗传转化体系,进一步阐明OfSAUR21基因与乙烯合成及信号传导通路之间的相互作用机制,为揭示桂花花瓣衰老的分子调控网络提供更深入的理论依据,同时也为通过基因工程手段调控桂花花期、延长观赏寿命提供潜在的应用靶点。
-
本研究从桂花中克隆得到1条SAUR同源基因,命名为OfSAUR21。OfSAUR21基因编码区长294 bp,编码97个氨基酸,序列包含完整的Auxin_inducible superfamily保守结构域。不同开花时期的基因表达水平表明该基因在花瓣开花后期表达最高,推测其可能与花瓣衰老有关。将OfSAUR21基因通过瞬时转化侵染到桂花花瓣中,RT-qPCR与代谢物分析确认发现:成功导入目的基因的桂花花瓣乙烯的释放量增加。因此,OfSAUR21基因在促进乙烯生成及调控桂花衰老中起着正调节功能。
Cloning and functional verification of OfSAUR21 gene in Osmanthus fragrans
doi: 10.11833/j.issn.2095-0756.20250446
- Received Date: 2025-08-21
- Accepted Date: 2025-09-28
- Rev Recd Date: 2025-09-24
- Available Online: 2025-10-29
- Publish Date: 2025-10-20
-
Key words:
- Osmanthus fragrans /
- SAUR gene /
- ethylene /
- gene clone /
- functional verification
Abstract:
| Citation: | GUO Hannian, XIONG Xin, HU Feiyang, et al. Cloning and functional verification of OfSAUR21 gene in Osmanthus fragrans[J]. Journal of Zhejiang A&F University, 2025, 42(5): 1059−1067 doi: 10.11833/j.issn.2095-0756.20250446 |












 DownLoad:
DownLoad: