-
植物内生固氮菌是指定殖在健康植物体内、与宿主植物进行联合固氮活动的一类原核微生物,是植物微生态系统的天然组分[1]。研究发现[2]:因为存在固氮菌,某些亚热带地区稻田可以连续百年不施氮肥而土壤氮水平不下降。固氮微生物一般分为3类:自生固氮菌、与植物共生固氮的微生物、与植物联合固氮的微生物。共生固氮微生物常见于豆科Leguminosae植物中[3],其固氮机制、菌肥开发等研究已相当成熟。联合固氮微生物最早由DÖBEREINER [4]从热带禾木科Gramineae牧草雀裨Paspalum thunbergii根际中分离获得,这种被命名为雀裨固氮菌Azotobacter paspali的微生物被认为是存在于根际的自由生活的固氮菌。它们可以部分入侵植物表层组织,依靠根系分泌物生存繁殖,不与宿主形成特异分化结构,因而固氮能力不及共生固氮菌,但这类固氮菌与植物关系密切。BARRAQUIO等[5]通过建立有效的水稻Oryza sativa内生固氮菌分离方法,测得水稻内生固氮菌数量为105~108 CFU·g−1;袁梅等[6]从湖南桂阳农田水稻分离出19株内生固氮菌,其中13株可抑制水稻苗期对镉的吸收;谭志远等[7]从青香茅Cymbopogon caesius与五节芒Miscanthus floridulus中分离出66株内生固氮菌共10个类群;BALDANI等[8]发现玉米Zea mays茎内存在内生固氮菌肺炎克雷伯氏菌Klebsiella pneumoniae,后者具有促进植物生长的作用。作为同属禾本科的木本植物,竹子内生固氮菌的报道较少。竹子是喜氮植物,对氮素要求较高;调查发现许多自然竹林没有氮肥补充但生长良好,提示竹子可能存在某种固氮菌,且后者具有较强的固氮潜能。顾小平等[9]从浙江淡竹Phyllostachys glauca根际分离到3株活性较高的固氮菌,从毛竹Phyllostachys edulis根表面分离到36株固氮菌,分别为多黏芽孢杆菌Bacillus polymyxa和地衣芽孢杆菌Baclicus lincheniformis [10];侯伟等[11]从簕竹Bambusa blumeana中分离到40株高效固氮酶活性菌株,分别属固氮螺菌属Azosprillum、大肠埃希氏菌属Escherichias、绿脓杆菌属Pseudomonas和水螺旋菌属Aquaspirillum;同时从簕竹[12]中分离到5株内生固氮菌,具有活性高、生长强势、可以通过自身代谢作用来适应环境酸碱性变化等特征。目前,国内关于竹子内生固氮菌的研究甚少。本研究以毛竹 (散生)、孝顺竹Bambusa multiplex (丛生)和狭叶青苦竹Pleioblastus chino (混生)为研究对象,利用分子生物技术研究不同生长型竹子根部及叶部的内生固氮菌、定殖土壤固氮菌的多样性和结构特征,探究不同竹种和不同器官间内生固氮菌多样性,探讨植株与定殖土壤内固氮菌多样性的关系,为竹子培育管理提供指导,也为开发有潜力的高效固氮菌,减少化学氮肥施用量、解决因化肥使用过度所造成的环境污染问题提供参考。
HTML
-
狭叶青苦竹、孝顺竹栽植于浙江农林大学竹种园内,毛竹栽植于浙江农林大学后山。所有林相均为纯林,所有土壤均为红黄壤。采集竹叶、细根及不同竹种根部定植土壤。被选植株要求生长力旺盛,远离边缘地带,未受到病虫害感染。每种竹子选取3株间隔约15 m的不同植株作为重复。土壤样品采自所选竹子根周,深度为0~30 cm。所有样品放入4 ℃冰箱保存备用。
-
经风干、磨细、过筛后,测定土样土壤化学性质。具体参考鲍士旦[13]的方法,测定土壤含水量、pH、有机质、有效氮、速效磷、速效钾等指标。由表1可知:3种竹子定植土壤的pH、有机质、有效氮、速效钾、速效磷等指标均未表现出显著性差异(P>0.05),说明土壤样品的化学性质差异不影响固氮菌种类。
种类 pH 有机质/(g·kg−1) 有效氮/(mg·kg−1) 速效磷/(mg·kg−1) 速效钾/(mg·kg−1) 毛竹 4.32±0.24 a 33.01±7.04 a 58.20±16.46 a 5.40±0.85 a 79.00±8.80 a 孝顺竹 4.64±0.27 a 36.85±10.81 a 66.14±14.46 a 4.99±0.61 a 69.33±17.02 a 狭叶青苦竹 4.72±0.54 a 37.67±9.16 a 57.34±15.63 a 4.65±0.94 a 70.67±4.50 a 说明:同列相同字母表示差异不显著(P>0.05) Table 1. Basic chemical properties of three soil
-
竹子叶与根的预处理参考谭致远等[7]方法。叶与根灭菌消毒后,剪碎放入无菌研钵。用液氮充分研磨破碎植物样品,参考高志民等[14]改良后的CTAB法提取植物内生微生物DNA,−40 ℃冰箱保存备用。
-
土壤样品过20目筛,按Mobio土壤微生物DNA试剂盒(Powersoil kit,美国)提取土壤微生物DNA。
-
固氮酶nifH基因采用聚合酶链式反应方法(PCR)扩增。第1轮正向引物FGPH19:5′-TACGGCAARGGTGGNATHG-3′,反向引物polR:5′-ATSGCATCATYTCRCCGGA-3′;第2轮正向引物AQER:5′-GACGATGTAGATYTCCTG-3′,反向引物polF-GC:5′-TGCGAYCCSAARGCBGACTC-3′。扩增程序参考DIALLO等[15]。
-
使用DCodeTM Universal Mutation Detection System (Bio-Rad,美国)对PCR产物进行变性梯度凝胶电泳,用GelDocTM EQ凝胶成像系统 (Bio-Rad,美国)拍照保存。
-
用polF(不含GC)与AQER引物扩增各竹种叶部、根部DNA,PCR产物用纯化试剂盒(Takara)纯化后与pEASY-T3 Vector(全金式生物技术公司,中国,北京)连接,转化到Trans1-T1感受态细胞中,涂在含有X-Gal、IPTG、氨苄青霉素的LB琼脂平板上进行蓝白斑筛选,用菌落PCR方法检测阳性克隆子。阳性克隆菌液送上海生工生物技术公司进行测序,序列通过NCBI进行Blast比对分析,获取相近典型菌株序列[16]。
-
利用Quantity one软件对DGGE电泳图进行指纹图谱分析,根据条带对比结果,通过未加权算术平均法进行聚类分析;计算Shannon多样性指数,使用SPSS软件进行单因素方差分析与两两比较(Ducan法);使用Mega 5.0,采用邻接法进行系统发育树分析。
1.1. 样品采集
1.2. 土壤基本化学性质测定
1.3. 竹子内生微生物DNA提取
1.4. 土壤微生物DNA提取
1.5. 固氮酶nifH基因扩增
1.6. 固氮酶nifH基因的变性梯度凝胶电泳(DGGE)
1.7. 固氮酶nifH基因的随机克隆
1.8. 数据分析
-
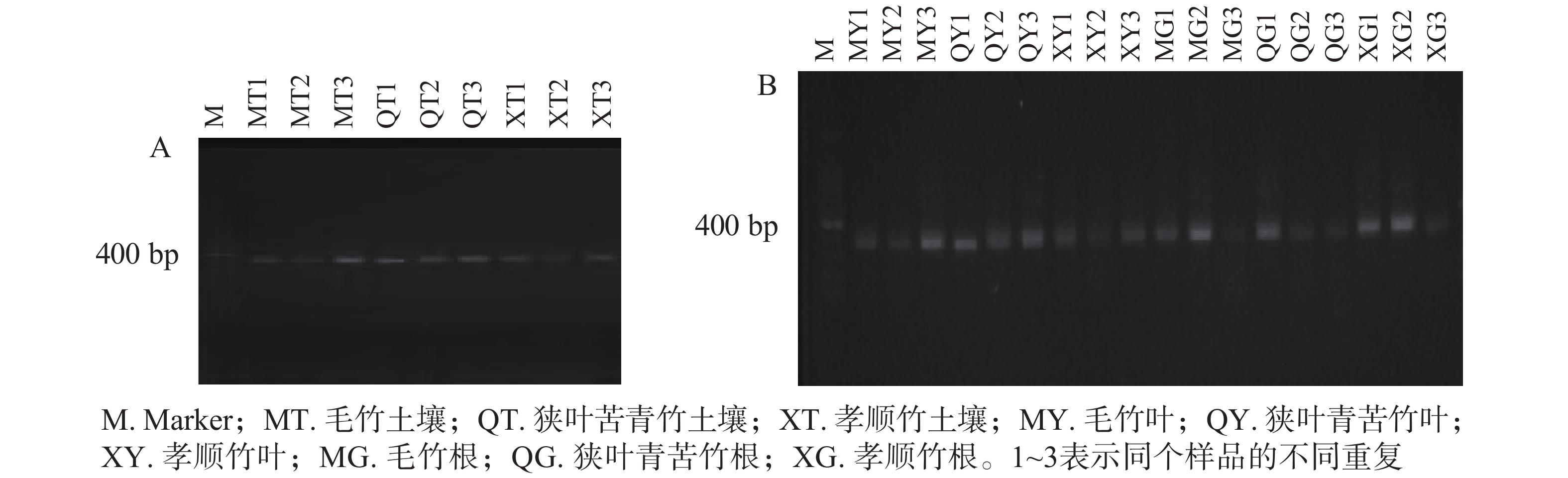
对土壤微生物、植物内生微生物DNA分别进行2轮巢氏PCR,获得土壤和植物固氮菌的nifH基因。由图1A和图1B可见:400~500 bp出现了一系列较为明亮的条带,即为目标nifH基因。
-
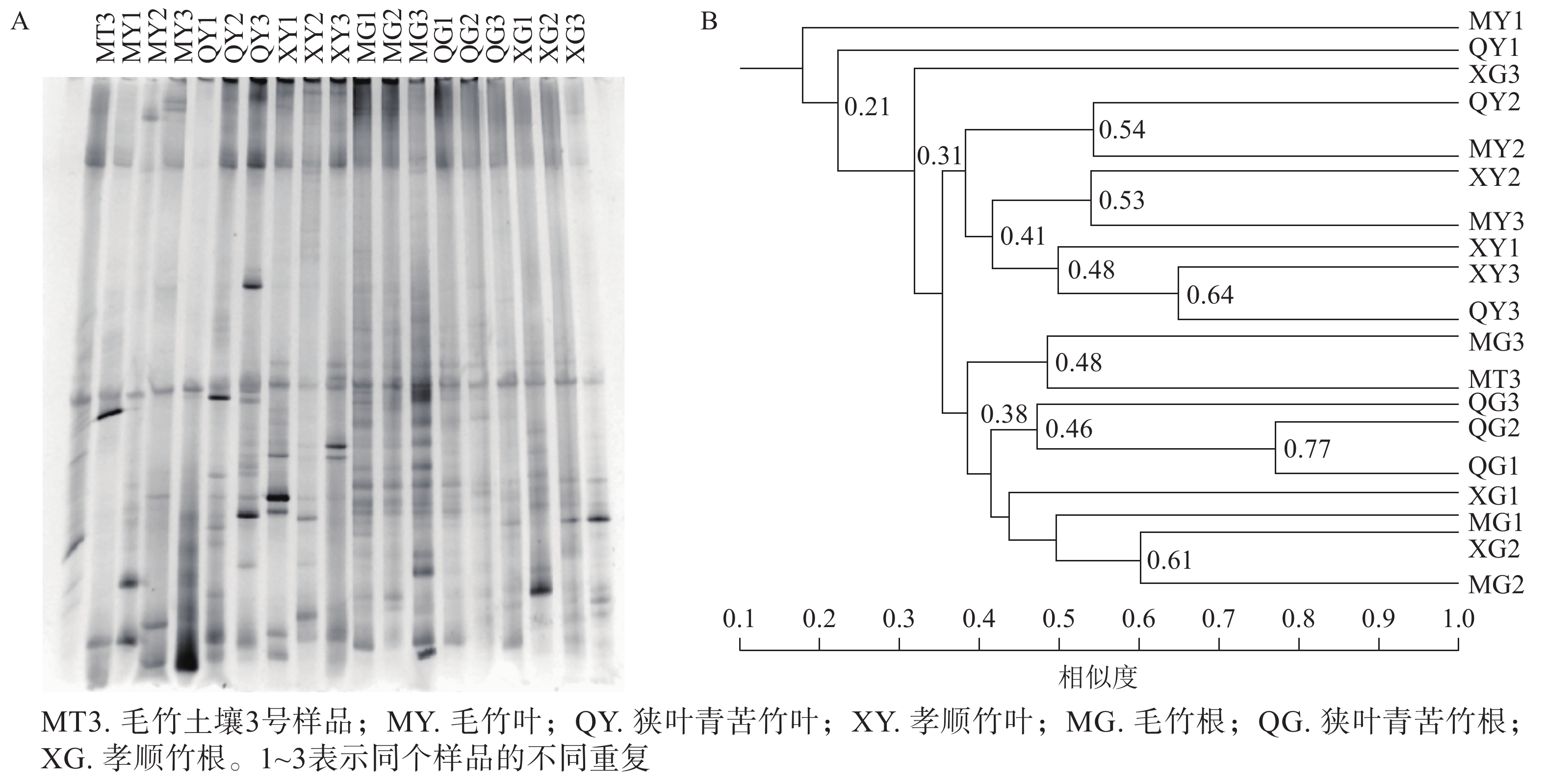
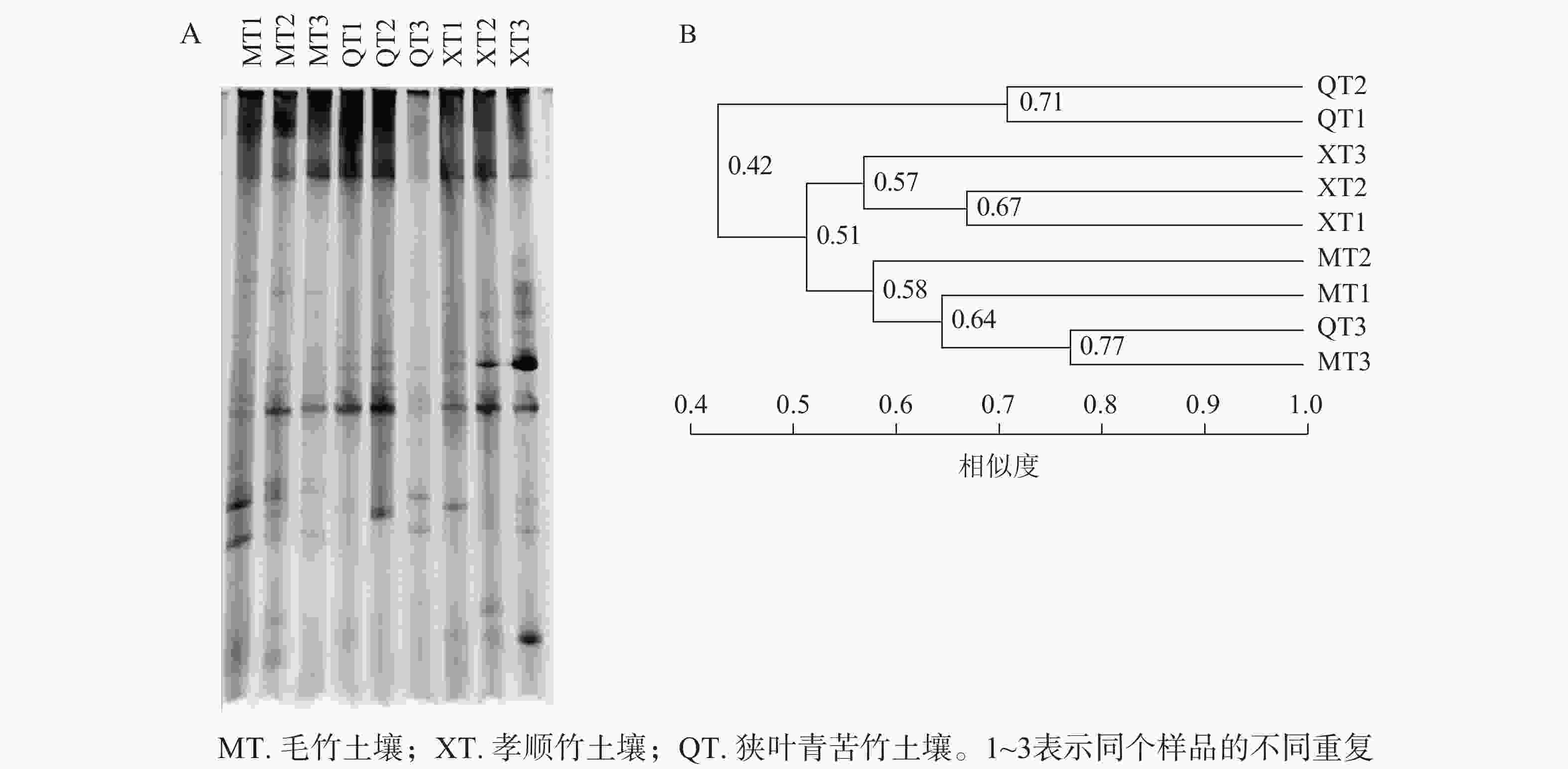
DGGE图谱中,条带数量代表样品所含固氮菌种类、多样性的丰富程度,条带粗细代表该种菌的生物量的大小(图2A)。使用Quantity one软件识别DGGE图中条带信息,以毛竹土壤3号为基准进行相似度分析,共识别出17个条带,且不同土壤样品之间出现一定数量的共有条带。这表明不同竹子定植的土壤存在着一定数量相同种类的固氮菌。
聚类分析中各样品之间的分支点数值代表植物内生固氮微生物之间亲缘关系的远近,数值越大越相似。由图2B可知:不同竹种之间、同一竹种重复样品间土壤固氮菌相似度不高。一般认为相似度达到0.60及以上时说明群体间相似性好[17]。本研究中孝顺竹3个土壤样品重复聚在一起,但整体相似度也仅为0.57,说明土壤中固氮菌群落的变异性较大。对于土壤微生物而言,复杂的物质组成和空间结构导致微生物的变异性增加。虽然3种土壤的理化性质没有显著差异(表1),但土壤的固氮菌仍然存在较大差异。
-
不同竹种、不同器官的内生固氮菌nifH基因DGGE指纹图谱如图3A所示。以毛竹土壤3号为参照,毛竹叶与根、狭叶青苦竹叶、孝顺竹叶与根等9个样品出现了明亮且粗大的条带,这些条带丰度较大,为优势固氮菌种。研究发现[18]:不同菌在组织内占有不同生态位,它们相互作用建立一种生态平衡,分离频率高的为植物体内数量大的优势种群,相反则为稀有种群。图3中,竹根的条带数明显多于竹叶,说明根部内生固氮菌种类多于叶部。刘丽辉等[19]发现普通野生稻Oryza rufipogon内生固氮菌在水稻根部分布最多,茎叶中偏少,呈自下而上逐级递减规律,与本研究一致。且竹根部内生固氮菌种类远多于土壤,说明竹子根部固氮菌不仅来源于土壤,还通过其他途径进入植株,如KLUEPFEL等[20]认为细菌是通过植物自然开口(水孔、气孔、皮孔和蜜腺等)和伤口(根磨损、动物摄食及收割多年植物等)进入植物体内的。
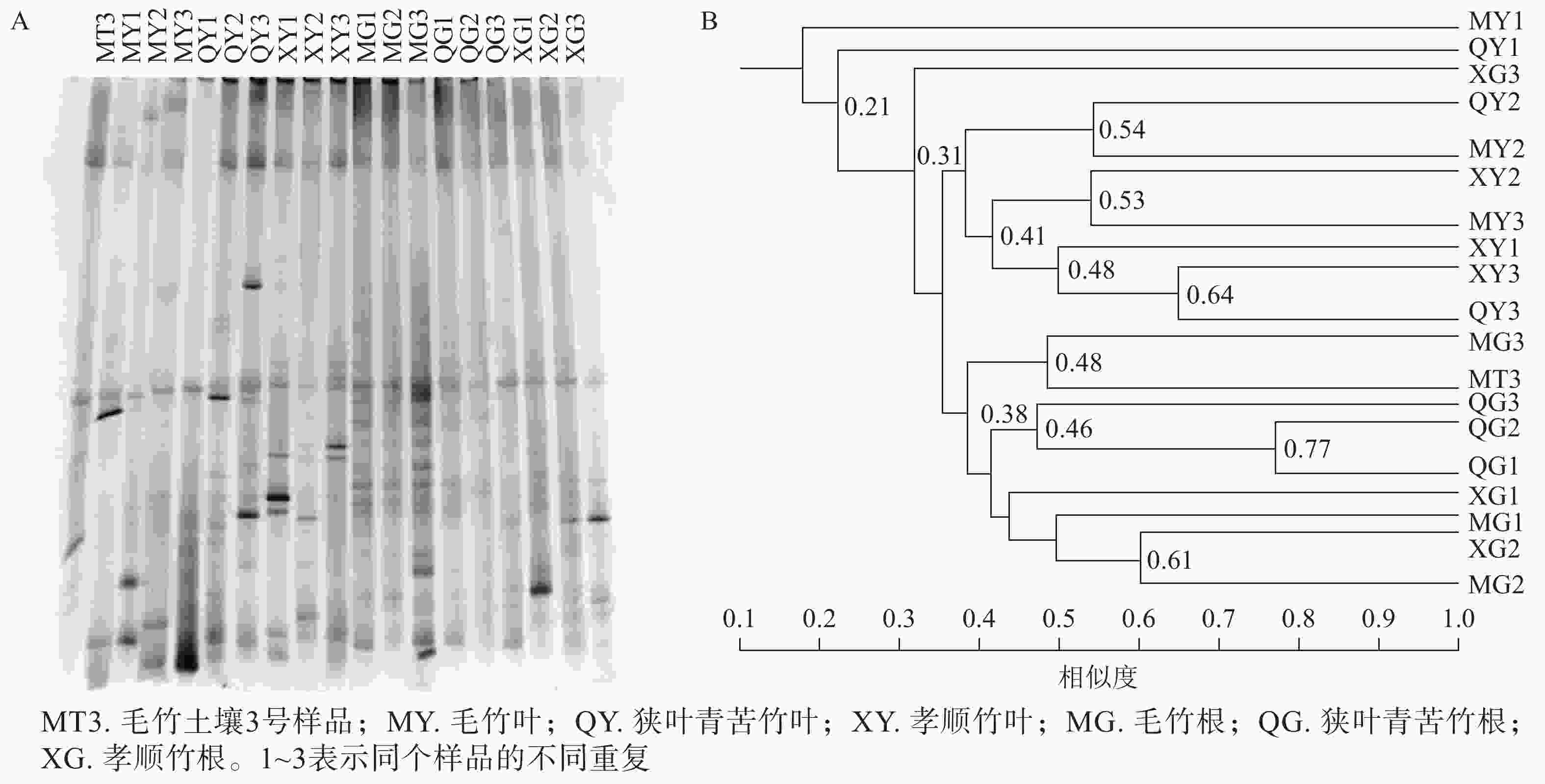
Figure 3. DGGE fingerprint of nifH gene endophytic diazotroph(A) and clustering analysis tree (B) in bamboo’s leaves and roots
聚类分析结果(图3B)表明:不同植物样品整体相似度不高,但大部分叶与根的样品明显区分,竹子叶与根分别聚在不同组,表明竹子器官差异是影响内生固氮定殖的重要因素。毛竹土壤样品(MT3)和根部样品聚集,但相似值仅有0.48,进一步说明了植株内生固氮菌仅有部分通过土壤进入,且大多定殖在根部,少部分扩散到叶部。以相似度大于0.60为群体间相似性很好的标准判断[17],同一品种同部位重复样品间的相似度均不高,除狭叶苦青竹1号与狭叶苦青竹2相似值达0.77外,其他样品3个重复都未能聚集,说明内生固氮菌种类与植物种类关系不大,遗传因子没有起到决定性作用。植物内生固氮菌的捕获具有一定的偶然性。
-
以指纹图谱信息为依据计算各个样品的Shannon多样性指数可以评价群落多样性。如表2所示:狭叶青苦竹根部内生固氮菌多样性最为丰富,显著高于叶部与土壤(P<0.05)。毛竹固氮菌多样性依次为根部(2.98)、土壤(2.75)、叶部(2.30),毛竹根部和土壤间无显著差异,但两者显著大于毛竹叶部(P<0.05),说明土壤固氮菌与根部内生固氮菌多样性都比叶部要丰富。孝顺竹叶部、根部内生固氮菌、土壤固氮菌间多样性差异不显著(P>0.05)。不同竹种不同部位固氮菌多样性不同,狭叶青苦竹和毛竹以叶部最低,孝顺竹叶部最高,这可能与竹子的生长繁殖方法有关;孝顺竹为丛生竹,竹子在某一个固定位置生长更新,根部和土壤中的固氮菌容易进入叶子;毛竹为散生竹,狭叶青苦竹虽然是混合型竹子,但以散生的居多,随着地下鞭的延伸更新,这些竹子不断改变着生地点,其本身的固氮菌可能也少,通过地下鞭和地上部分的通道到达叶子中的固氮菌便更少。
种类 叶部 根部 土壤 狭叶青苦竹 2.43±0.30 Bb 2.89±0.12 ABa 2.36±0.09 Bb 毛竹 2.30±0.28 Bb 2.98±0.19 Aa 2.75±0.17 Aa 孝顺竹 2.73±0.10 Aa 2.53±0.26 Ba 2.70±0.15 Aa 说明:大写字母表示同列间差异显著(P<0.05),小写字母 表示同行间差异显著(P<0.05) Table 2. Shannon indexes of the three bamboos’ leaves, roots and soil
对不同竹种相同部位内生菌多样性分析发现:孝顺竹叶部内生菌多样性显著高于狭叶青苦竹和毛竹(P<0.05),根部则显著低于狭叶青苦竹和毛竹(P<0.05);毛竹和孝顺竹土壤固氮菌多样性显著高于狭叶青苦竹(P<0.05);说明对于毛竹与狭叶青苦竹而言,固氮菌群落更偏爱在根部定殖,对于孝顺竹而言,固氮菌群落更喜爱在叶内定殖。
-
对不同竹种根部/叶部的内生固氮菌DNA作随机克隆并测序(表3),在培养基上共得到484个蓝色斑点,经验阳后送测121个,序列经BLAST数据库对比,得到83株菌株,其中12株为可培养固氮菌,归属于8种菌属,其余71株为不可培养固氮菌(来源于毛竹叶8株,毛竹根14株,孝顺竹叶7株,孝顺竹根14株,狭叶青苦竹叶8株,狭叶青苦竹竹根20株),尚未被命名,不可培养的比例高达86%;推测原因可能是不可培养固氮菌在竹子内生固氮菌中处于优势地位,也可能是竹子中某些可培养的内生固氮菌尚未在其他植物中被分离鉴定,这也进一步说明了该类固氮菌的特殊性。12株可培养的固氮菌大部分为根组织内生菌(表4),除Immundisolibacter和红长命菌属Rubrivivax外,均已报道具有固氮基因[19,21-28]。8个菌属中,慢生根瘤菌属Bradyrhizobium出现频率最高,一般是从大豆Glycine max根部分离得到,如XU等[21]从大豆和野大豆Glycine soja根瘤中分离出了与大豆结瘤的超慢生辽宁大豆根瘤菌Bradyrhizobium liaoninggense。固氮螺菌属Azospirillum具有明显的固氮功能,在植物根、茎、叶中广泛存在[19];此外,翟超男[22],周建娇[23]分别从海滨雀稗Paspalum vaginatum和白菜Brassica pekinensis根发现具有固氮功能的肠杆菌Enterobacter sp.;李洁琼等[24]从玉米根际中分离出具有促生作用的Kosakonia radicincitans strain YD4同样具有固氮功能,而Kosakonia是从肠杆菌属Enterobacter中分离出来的新属。TAN等[25]通过南方杂交法与乙炔还原法发现Dechlorosoma suillum中具有nifH基因;唐凯等[26]在研究浑善达克沙地苔藓Bryophyta结皮下层土壤细菌时发现Azohydromonas具有固氮功能。红长命菌可以氨为氮源,是目前发现的唯一能分泌蛋白酶的光合细菌[27],Immundisolibacter是变形杆菌的一种新细菌,具有降解多环芳香烃的功能[28]。本研究从毛竹根部和孝顺竹根部鉴定出的红长命菌和Immundisolibacter中存在固氮基因,推测此2种菌很可能是具有固氮能力的新种。
样品来源 相似菌种基因 毛竹叶 HP-46 nitrogenase (nifH) gene、HP-76 nitrogenase (nifH) gene、nifH gene for nitrogenase iron protein、nitrogen-
fixingbacteriumcloneb1 -HA3-1nitrogenase iron protein (nifH) gene、isolate DGGE gel band 5 nitrogenase iron protein (nifH)
gene、J19I(16) nitrogenase (nifH) gene、HP-12 nitrogenase (nifH) gene、TNO_elv_99f12 dinitrogenase reductase (nifH) gene毛竹根 nifH gene for nitrogenase iron protein、HP-27 nitrogenase (nifH) gene、HP-84 nitrogenase
(nifH) gene、HP-84 nitrogenase (nifH) gene、CI(12) nitrogenase (nifH) gene、HP-14 nitrogenase (nifH) gene、TII(5) nitrogenase
(nifH) gene、HP-48 nitrogenase (nifH) gene、HP-75 nitrogenase (nifH) gene、J19I(12) nitrogenase (nifH) gene、TNO_elv_81h1
dinitrogenase reductase (nifH) gene、TNO_elv_81h1 dinitrogenase reductase (nifH) gene、HP-46 nitrogenase (nifH) gene、HP-75
nitrogenase (nifH) gene孝顺竹叶 HP-215 nitrogenase (nifH) gene、HP-5 nitrogenase (nifH) gene、HP-218 nitrogenase (nifH) gene、CY1-01 nitrogenase iron
protein (nifH) gene、HP-56 nitrogenase (nifH) gene、HP-226 nitrogenase (nifH) gene、HP-157 nitrogenase (nifH) gene孝顺竹根 isolate DGGE gel band 18 nitrogenase iron protein (nifH) gene、isolate DGGE gel band 12 nitrogenase iron protein (nifH) gene、
HP-226 nitrogenase (nifH) gene、partial nifH gene for dinitrogenase reductase、HP-90 nitrogenase (nifH) gene、partial nifH
gene for dinitrogenase reductase、HP-237 nitrogenase (nifH) gene、HP-246 nitrogenase (nifH) gene、HP-246 nitrogenase
(nifH) gene、HP-51 nitrogenase (nifH) gene、C13L7NH26 dinitrogenase reductase (nifH) gene、isolate DGGE gel band 20
nitrogenase iron protein (nifH) gene、b1-HA3-1 nitrogenase iron protein (nifH) gene、431 nitrogenase iron protein (nifH) gene狭叶青苦竹叶 TC30 NifH (nifH) gene、C12L6CK9 dinitrogenase reductase (nifH) gene、C13L6NH2 dinitrogenase reductase (nifH) gene、
C12L7NH35 dinitrogenase reductase (nifH) gene、nifH gene for nitrogenase iron protein(partial cds)、C13L10NH15
dinitrogenase reductase (nifH) gene、C13L8CK17 dinitrogenase reductase (nifH) gene、HP-75 nitrogenase (nifH) gene狭叶青苦竹根 isolate DGGE gel band 14 nitrogenase iron protein (nifH) gene、Rb3_NifH_48 nitrogenase gene、C12L8CK4 dinitrogenase
reductase (nifH) gene、WK-A5 nitrogenase iron protein (nifH) gene、486 nitrogenase iron protein (nifH) gene、C13L8NL22
dinitrogenase reductase (nifH) gene、C13L6CK40 dinitrogenase reductase (nifH) gene、99 nitrogenase iron protein (nifH)
gene、HP-201 nitrogenase (nifH) gene、HP-2 nitrogenase (nifH) gene、243 nitrogenase iron protein (nifH) gene、
C12L10NL20 dinitrogenase reductase (nifH) gene、C13L10NL22 dinitrogenase reductase (nifH) gene、isolate DGGE gel band
11 nitrogenase iron protein (nifH) gene、C12L7CK25 dinitrogenase reductase (nifH) gene、HP-159 nitrogenase (nifH) gene、
HP-215 nitrogenase (nifH) gene、HP-90 nitrogenase (nifH) gene、C12L7NH35 dinitrogenase reductase (nifH) gene、
b1-HA3-1 nitrogenase iron protein (nifH) geneTable 3. Distribution of unculturable bacteria based on nifH gene (The similarity is more than 98%)
类群 样品来源 NCBI中最相近菌种(登录号) 序列相似性/% 慢生根瘤菌属 Bradyrhizobium MG38 Bradyrhizobium sp.(KC356360.1) 98 QG11 Bradyrhizobium sp.(AB079616.1) 99 QG255 Bradyrhizobium sp.(LT859959.1) 99 XG32 Bradyrhizobium sp.(CU234118.1) 99 Dechlorosoma MG31 Dechlorosoma suillum(CP003153.1) 98 固氮螺菌属 Azospirillum XG434 Azospirillum canadense strain(GU256446.1) 99 肠杆菌属 Enterobacter XY47 Enterobacter sp.(KC989924.1) 99 Kosakonia MG6 Kosakonia sacchari strain(KR075981.1) 99 XG52 Kosakonia sacchari strain(KR075959.1) 99 变形杆菌属 Gammaproteobacteria XG255 Immundisolibacter cernigliae strain (CP014671.1) 99 红长菌属 Rubrivivax MG16 Rubrivivax gelatinosus strain(KF800058.1) 98 Azohydromonas XG66 Azohydromonas australica(AB188121.1) 100 Table 4. Distribution of culturable bacteria of nifH gene
孙建光等[29]从小麦Triticum aestivum、水稻、白菜、玉米、芹菜Apium sp.等5种农作物中分离到25个属的内生固氮菌。本研究选取伯克霍尔德氏菌Burkholderia sp. strain STM678、假单胞菌Pseudomonas corrugata、芽孢杆菌Bacillus aryabhattai strain B8W22和克雷伯氏菌Klebsiella variicola等4个优势种作为参照,与3种竹子中筛选出的克隆序列构建系统发育树。由图4可见,除1株来源于狭叶青苦竹的菌株(不可培养)与伯克霍尔德氏菌聚在一起外,其他菌株均远离参照菌株。具体来看,慢生根瘤菌属位于发育树中部位置,红长命菌属、Kosakonia、肠杆菌属、脱氯菌属Dechlorosoma、固氮螺菌属亲缘关系接近,Immundisolibacter cernigliae位于发育树的偏顶端位置,亲缘关系远离以上5个属。牛艳芳[30]对内蒙古植被主要建群植物的根际固氮菌进行分离时发现:6个地区的固氮菌类群存在显著差异,有些固氮菌分布具有区域性。推测这种大区域环境差异造成的固氮菌类群差异,可能也是竹子内生固氮菌与其他作物内生固氮菌存在较大区别的原因。
2.1. 不同来源固氮菌PCR产物
2.2. 土壤固氮菌DGGE分析
2.3. 竹子根部与叶部内生固氮菌DGGE分析
2.4. 不同来源固氮菌Shannon多样性指数分析
2.5. 竹子根部与叶部内生固氮菌的随机克隆
-
不同竹种定植的土壤中存在着一定数量的同种固氮菌,但整体区系的相似度不高。同一竹种不同器官(根与叶)之间、不同竹种相同器官之间、土壤与植物之间的固氮菌群落结构差异显著;同种竹叶部或根部的重复样品相似度均不高。3种竹子根部内生固氮菌种类总体显著多于叶部,土壤固氮菌群落结构与根部内生固氮菌相似度高于叶部;不同竹种、不同器官之间固氮菌群落Shannon多样性指数差异显著,孝顺竹叶部与土壤大于根部,狭叶青苦竹和毛竹根部大于叶部和土壤。植物样品随机克隆测序BLAST对比得到83株固氮菌,其中12株为可培养固氮菌株,分别属于慢生根瘤菌属、Dechlorosoma、固氮螺菌属、肠杆菌属、Kosakonia、变形杆菌属、红长菌属、Azohydromonas等8个属,与已报道对竹子内生固氮菌研究相比,仅与侯伟等[12]从簕竹中分离得到了相同的固氮螺菌,但未发现其他相同种属。系统发育树结果表明3种竹子内生固氮菌显著不同于其他植物中分离得到的内生固氮菌。










 DownLoad:
DownLoad: