-
丹参Salvia miltiorrhiza为唇形科Lamiaceae鼠尾草属Salvia多年生草本植物,又称红根、赤参[1−2]。其主要药用部位为干燥根或根茎,具有活血通经、祛瘀止痛、清心除烦、凉血消痈等功效[3]。研究表明:丹参含有多种化学活性成分,主要为脂溶性的萜类化合物和水溶性的酚酸类成分,具有保护心肌、抗凝血、抗炎、抗肿瘤等药理学功效[4−5]。目前,丹参面临野生种质资源被破坏、栽培生长周期长、有效成分含量低等问题[6],使得丹参品质退化,其原料药在临床上的可持续利用受限。因此,研究丹参药用活性成分的生物合成分子机制,可为丹参品种的选育提供科学依据。
茉莉酸信号分子参与植物生长发育等多个生理活动的调控,尤其在生物和非生物胁迫的防御反应中发挥着关键作用[7−8]。GE等[9]研究表明:甲基茉莉酸(MeJA)能诱导促进丹参酮的合成。ZHOU等[10]研究发现:丹参毛状根用MeJA处理后,酚酸类成分的含量显著提高。迄今为止,丹参酮在生物合成途径上的相关合成酶基因被大量克隆与鉴定[11−14];多个具有调控功能的转录因子如bHLH、MYB等也相继被挖掘[15−21],这些研究为全面解析转录因子调控丹参次生代谢物生物合成奠定了良好的基础。研究表明:bHLH类转录因子能响应MeJA信号调控植物次生代谢物的合成[21−25]。本研究通过比较转录组数据库,筛选获得1个响应MeJA诱导显著上调的丹参bHLH类转录因子。采用同源克隆的方法,克隆了该转录因子,命名为SmJRB2基因。基于农杆菌Agrobacterium tumefaciens介导的丹参遗传转化技术,对SmJRB2基因在丹参药用成分代谢合成中的功能及其表达特征进行了分析,以探究SmJRB2基因在MeJA信号通路中的功能,为解析丹参品质的形成机制和品种分子选育提供理论支撑。
-
丹参无菌苗,亚细胞定位瞬时表达载体和遗传转化超表达载体(pHB-X-YFP),大肠埃希菌Escherichia coli DH5α,农杆菌菌株C58C1、GV3101和EHA105为浙江中医药大学中药生物技术实验室保存;平末端试剂盒(pEASY-Blunt Cloning,北京全式金生物科技公司),柱式质粒DNA小量抽提试剂盒(上海生工生物公司),植物总RNA提取试剂盒和实时荧光定量PCR (RT-qPCR)试剂盒(天根生物科技有限公司),限制性核酸内切酶(Thermo Fisher Scientific),PowerUpTM SYBRTM Green Master (Applied Biosystem)。
-
按照植物总RNA提取试剂盒操作说明,提取丹参总RNA,用质量浓度为1%的琼脂糖凝胶电泳来检测其完整性,并使用微量核酸蛋白测定仪测定RNA的浓度与波长260、280 nm下吸光度比值D(260/280),检测合格后备用。使用RT-qPCR试剂盒对检测合格的RNA进行反转录,合成双链cDNA备用。
-
以丹参cDNA为模板,基于已测得的丹参转录组数据(NCBI登录号:GSE100970)拼接获得的SmJRB2序列信息,设计特异性引物组合SmJRB2-BamHⅠ-F和SmJRB2-SpeⅠ-R,以及SmJRB2-Anti-BamHⅠ-F和SmJRB2-Anti-SpeⅠ-R (表1),用来扩增目标SmJRB2基因序列;将PCR扩增产物进行琼脂糖凝胶电泳检测,将获得的目的DNA片段切下,通过胶回收试剂盒回收;取5 μL胶回收产物进行凝胶电泳检测,剩余的DNA回收片段用于克隆载体pMD-19T构建、转化大肠杆菌、阳性克隆鉴定与测序检测。测序正确的SmJRB2基因序列用于pHB超表达和反义抑制表达载体的构建。
引物名称 引物序列 (5′→3′) 引物名称 引物序列 (5′→3′) SmJRB2-BamHⅠ-F TCTCTCTCTAAGCTTGGATCCATGGGAAAGAAAGT ATGGTGGAATGAAGAAG SmCPS1-609R
SmKSL1-1480FTTCGAACCCACAAGTCATGT
GTGTGACCCTTCTGCTAGCASmJRB2-SpeⅠ-R GCCCTTGCTCACCATACTAGTTTTCAAGAGAGCAG CAGTTAACTTATCTTTCA SmKSL1-1630R
SmDXS2-1828FTGCATTGTCTTGGGAAGATG
TTGGAGATTGGGAAGGGAAGGATSmJRB2-Anti-BamHⅠ-F GGACTAGTATGGGAAAGAAAGTATGGTGGAATG SmDXS2-1980R
SmDXR-1248FAGGCTTGCAGAATCTCGCATCAG
CGACGAGAAAATCGGATACCTGGSmJRB2-Anti-SpeⅠ-R CGGGATCCTCATTTCAAGAGAGCAGCAGTTAACT SmDXR-1424R
SmHMGR-QFCATACAAGAGCAGGACTCGAACC
TCGTTTTCAATAAGTCGAGTAGAQF23 CCAAAGTTGTAAAGGCGTTGAGA SmHMGR-QR ATTCTGAAGGAAGTCCAAAACAT NOS-R TGGTGCAGATGAACTTCAGGGT SmCYP76AH1-1010F TCGTGGATGAGTCGGCAAT rolB-F GCTCTTGCAGTGCTAGATTT SmCYP76AH1-1168R TGAGTATCTGAGTTCCCT rolB-R GAAGGTGCAAGCTACCTCTC SmActin-QF1011 AGCACCGAGCAGCATGAAGATT SmCPS1-459F GATCGCCTCGTCAATACCAT SmActin-QR1210 AGCAAAGCAGCGAACGAAGAGT Table 1. Primer sequences used in this study
-
使用Vector NTI对候选基因的氨基酸进行同源比对分析,比较其保守结构域并着色。通过网站NCBI的blastp工具筛选同源氨基酸序列,采用MEGA 5.1绘制进化树。使用网站ExPASy分析蛋白质的分子量和理论等电点;使用网站TMHMM Server v2.0分析其是否有跨膜结构域;使用网站SignalP 4.1 Server分析基因信号肽[26−28]。
-
向培养1个月且生长状态良好的毛状根中加入10 µmol·L−1的MeJA进行诱导处理,选择0、0.5、1.0、2.0、4.0、6.0、9.0和12.0 h共8个时间点进行收获,每个时间点3次重复。以稀释20倍的cDNA为模板,根据需检测的基因设计特异RT-qPCR检测引物(表1)。以丹参Actin为内参基因,进行RT-qPCR检测,反应体系:8.0 μL RNase-free ddH2O,10.0 μL 2×SuperReal PreMix,0.2 μL 50×ROX Reference Dye,0.4 μL正向引物(QF),0.4 μL反向引物(QR),1.0 μL模板(cDNA)。
-
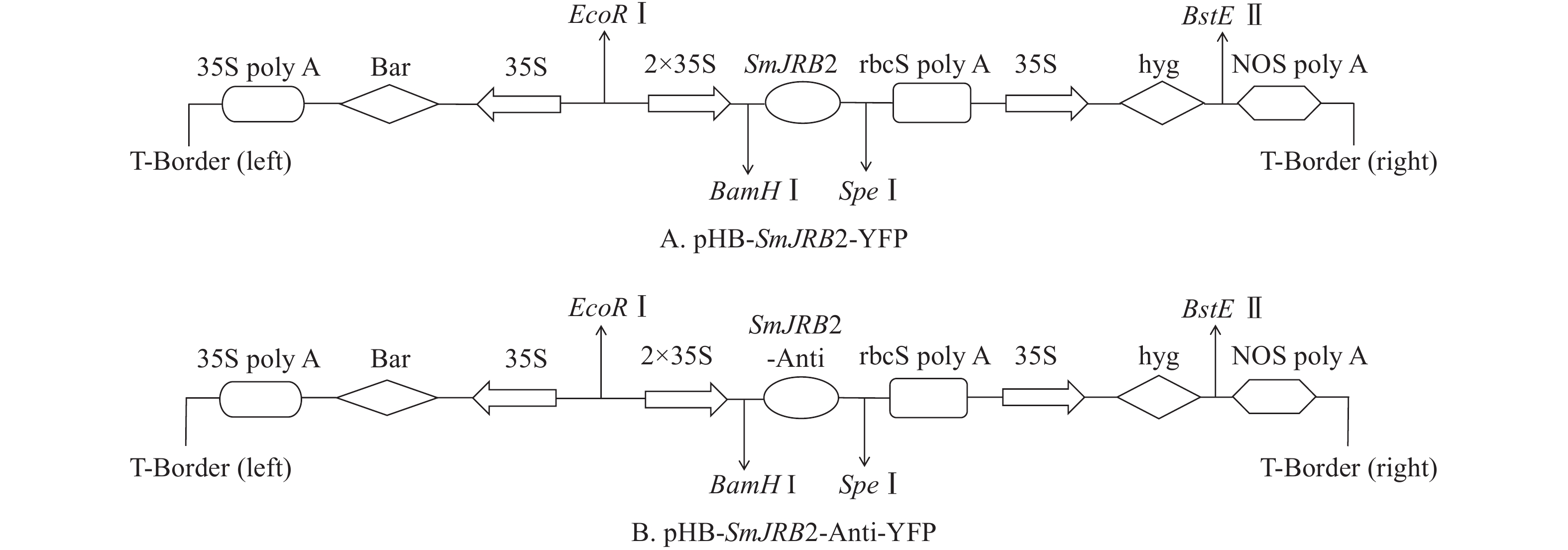
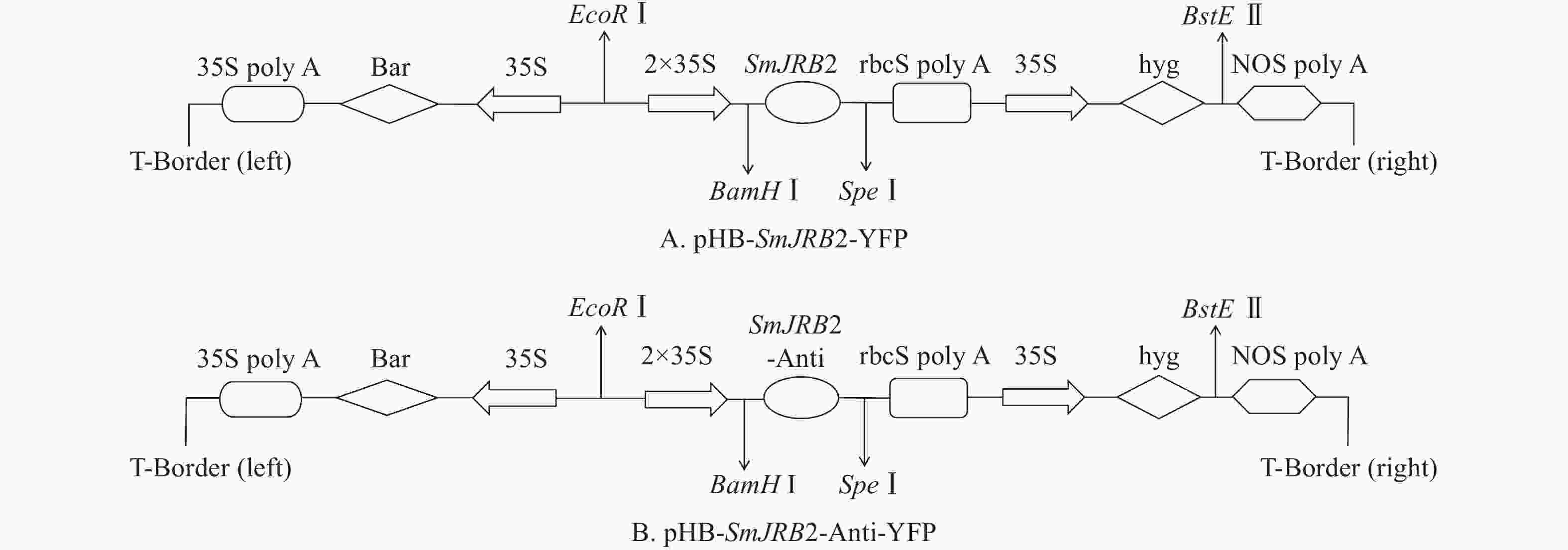
将上述测序正确的pMD-19T-SmJRB2大肠埃希菌阳性克隆以及包含pHB-X-YFP空质粒的大肠埃希菌,分别提取质粒后用BamHⅠ和SpeⅠ进行双酶切、电泳和切胶回收纯化;取SmJRB2基因回收片段与pHB-X-YFP空质粒双酶切回收的大片段进行连接、转化大肠埃希菌、阳性克隆鉴定及再转化C58C1农杆菌。
-
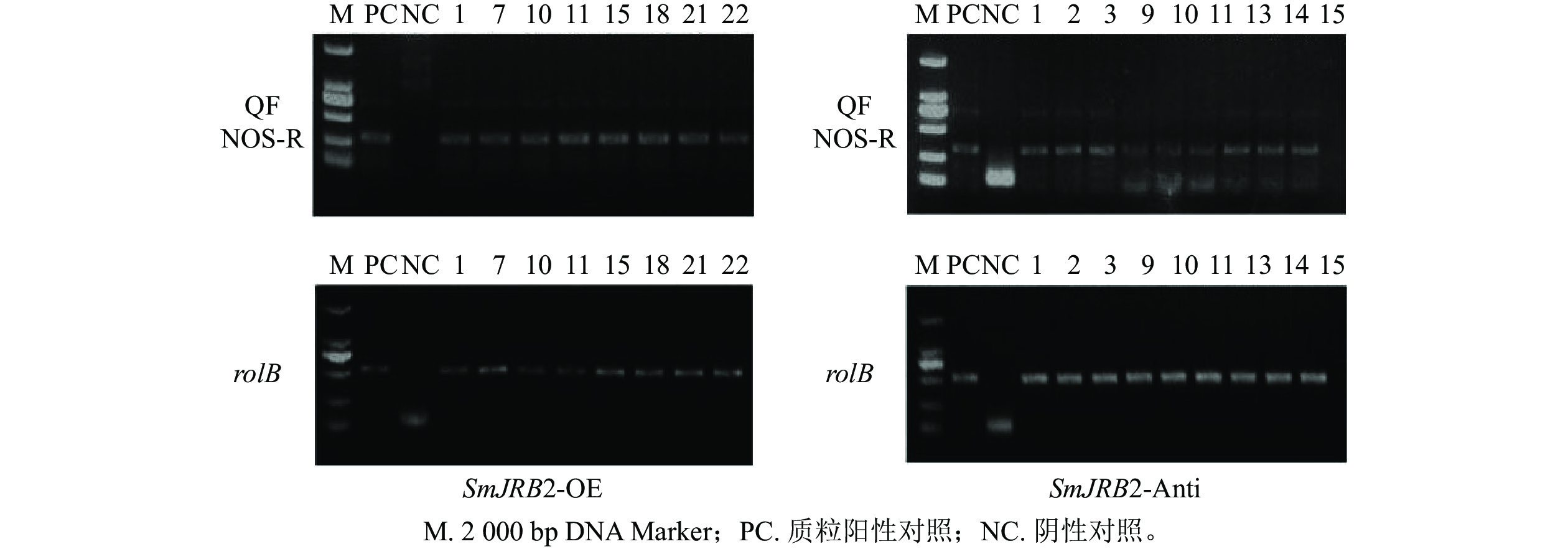
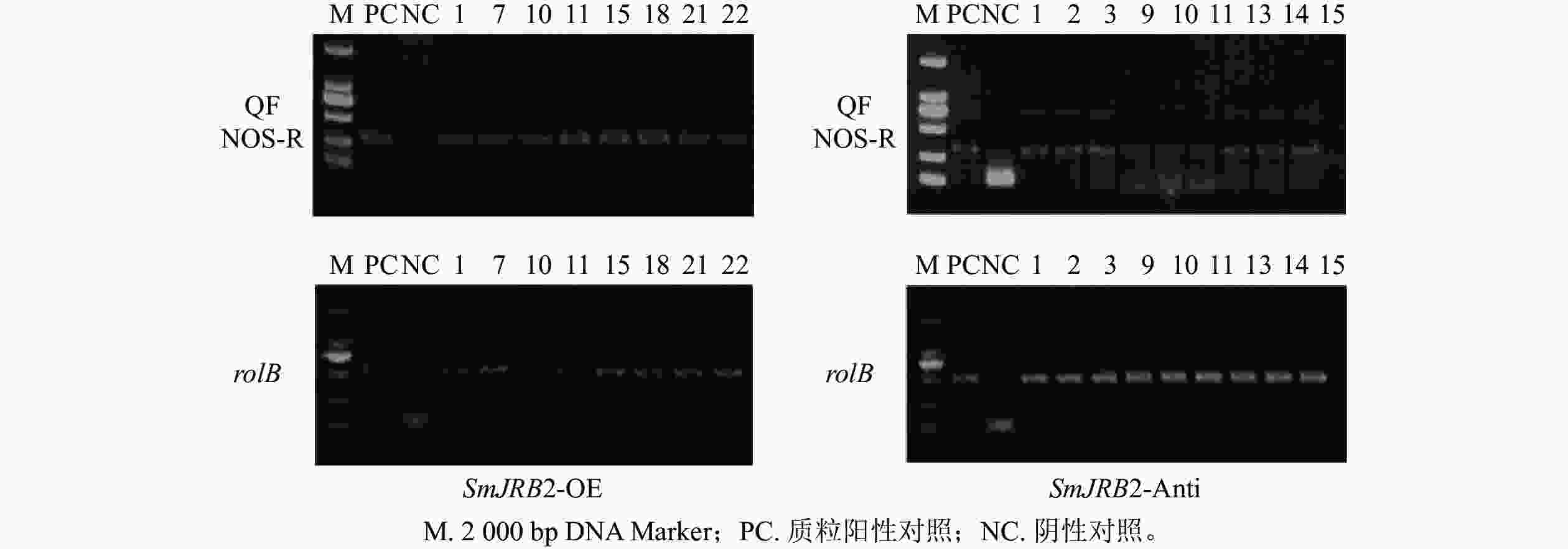
采用十六烷基三甲基溴化铵法(CTAB)法[13]分别提取上述获得的丹参毛状根系总DNA,作为PCR反应扩增的模板;再依据SmJRB2基因编码序列设计上游引物QF,pHB载体中NOS终止子设计下游引物NOS-R,扩增特异性DNA片段序列;再采用引物rolB-F和rolB-R扩增rolB基因(表1)。将不含SmJRB2基因的pHB空载体进行遗传转化,以获得超表达和反义抑制的毛状根,再以提取的总DNA为模版进行扩增,作为PCR鉴定的阴性对照。PCR反应条件如下:94 ℃预变性5 min;94 ℃变性45 s、58 ℃退火 45 s、72 ℃延伸1 min,35个循环;72 ℃延伸8 min。PCR扩增产物经琼脂糖凝胶电泳验证是否为阳性毛状根系。
-
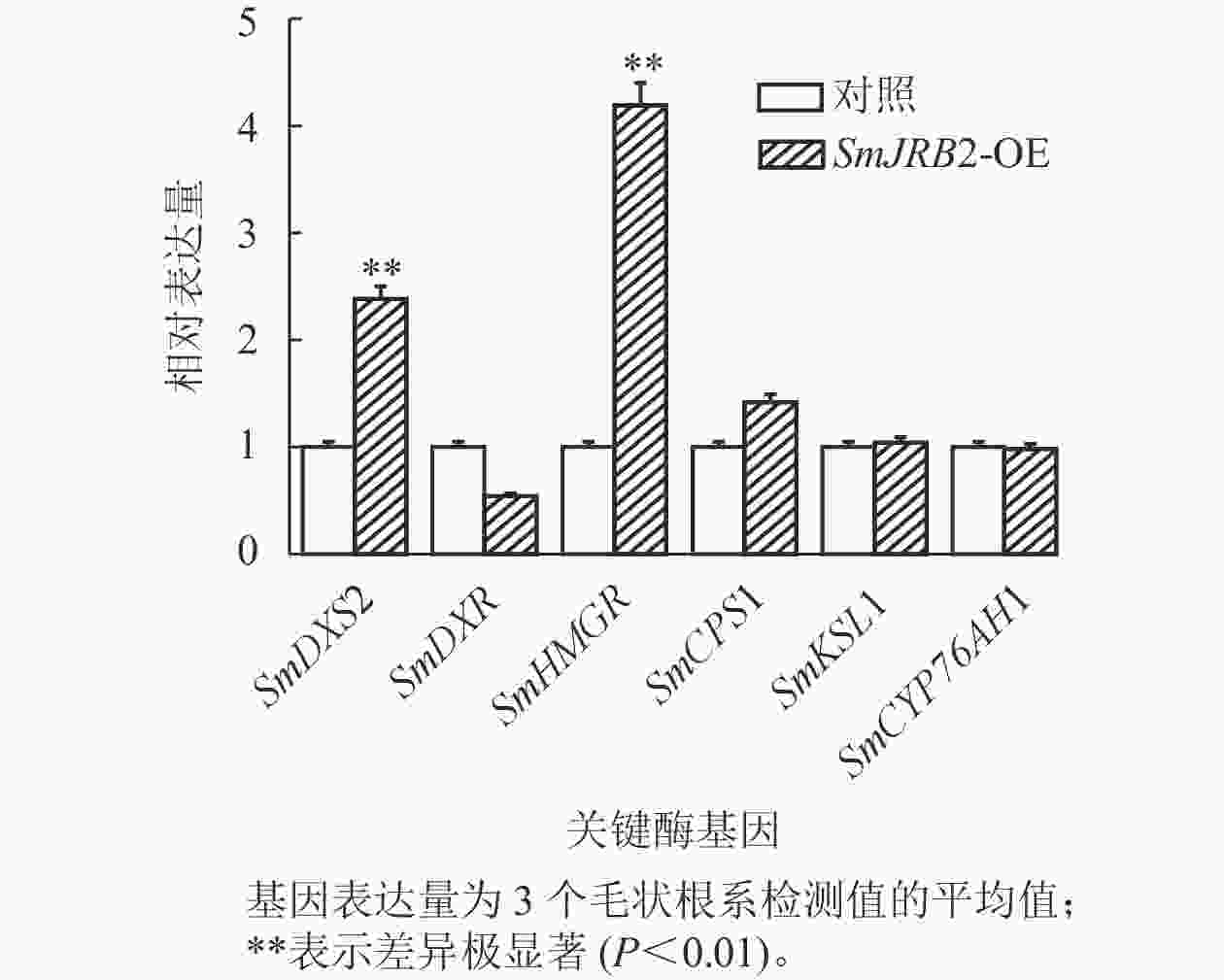
根据阳性鉴定结果,筛选出超表达效果较好的阳性毛状根,将生长良好的毛状根接入100 mL 1/2 MS液体培养基中,在摇床中25 ℃、120 r·min−1避光扩大培养,培养至60 d即可收根。分别检测丹参酮合成途径DXS2、DXR、HMGR、CPS1、KSL1和CYP76AH1基因的表达量,检测引物如表1所示。
-
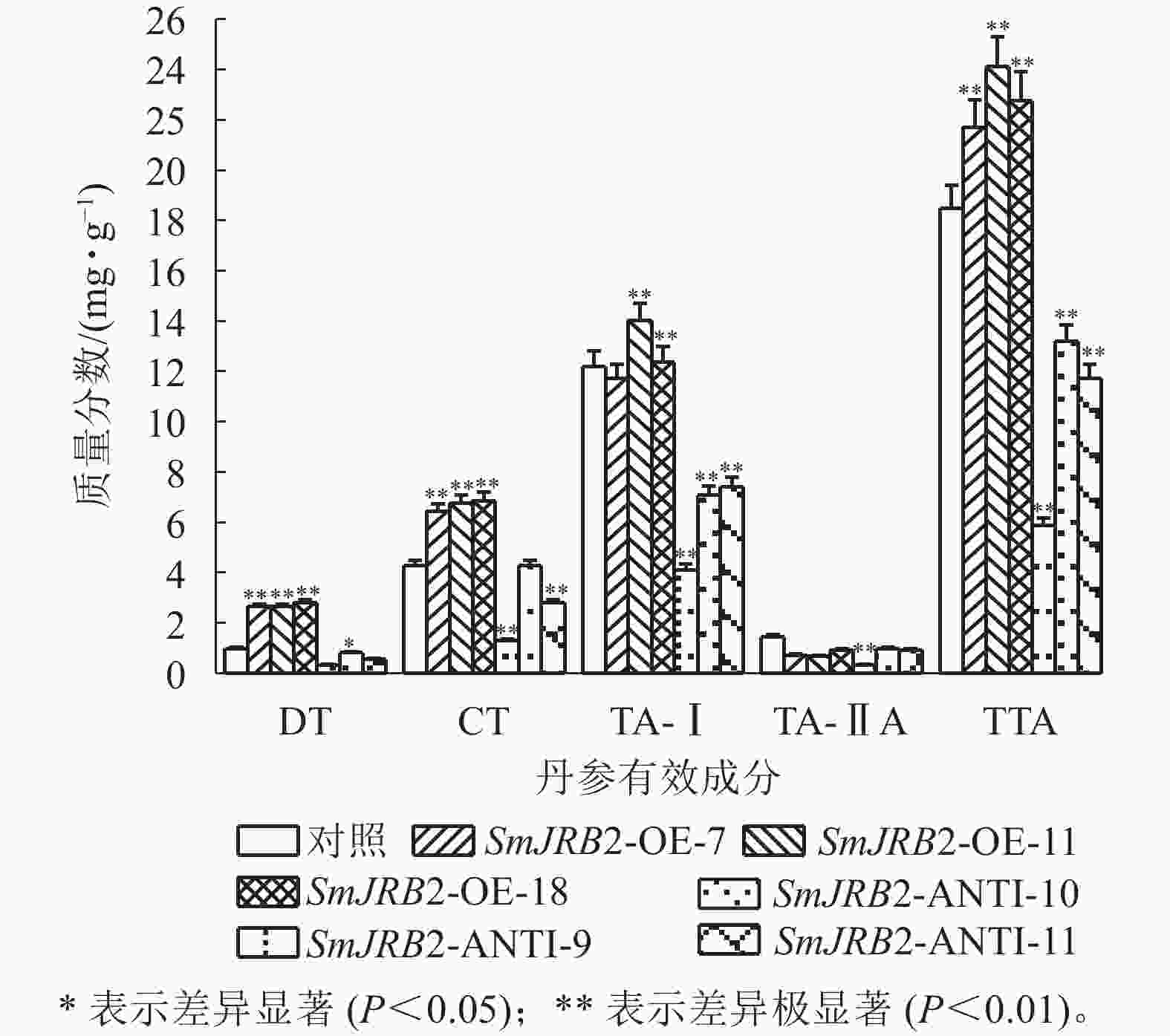
分别收集3个SmJRB2超表达与3个抑制表达毛状根系,用液氮研磨成粉,冷冻干燥处理24 h;取100 mg干粉,加入16 mL甲醇/三氯甲烷(体积比为3∶1),超声处理1 h后再过夜浸提,12 000×g离心10 min收集上清液倒入蒸馏烧瓶中,50 ℃蒸馏去掉提取液,6 500×g离心10 min收集药效提取物干物质。收集的干物质用1 mL蒸馏水充分溶解,再用0.22 µmol·L−1滤膜过滤。过滤后的样品通过高效液相色谱(HPLC)检测毛状根中丹参酮的质量分数[29]。
-
所有测试数据用SPSS 16.0进行t检验单因素方差分析。所有基因表达量和丹参酮质量分数检测均为3次生物学重复,取平均值。
-
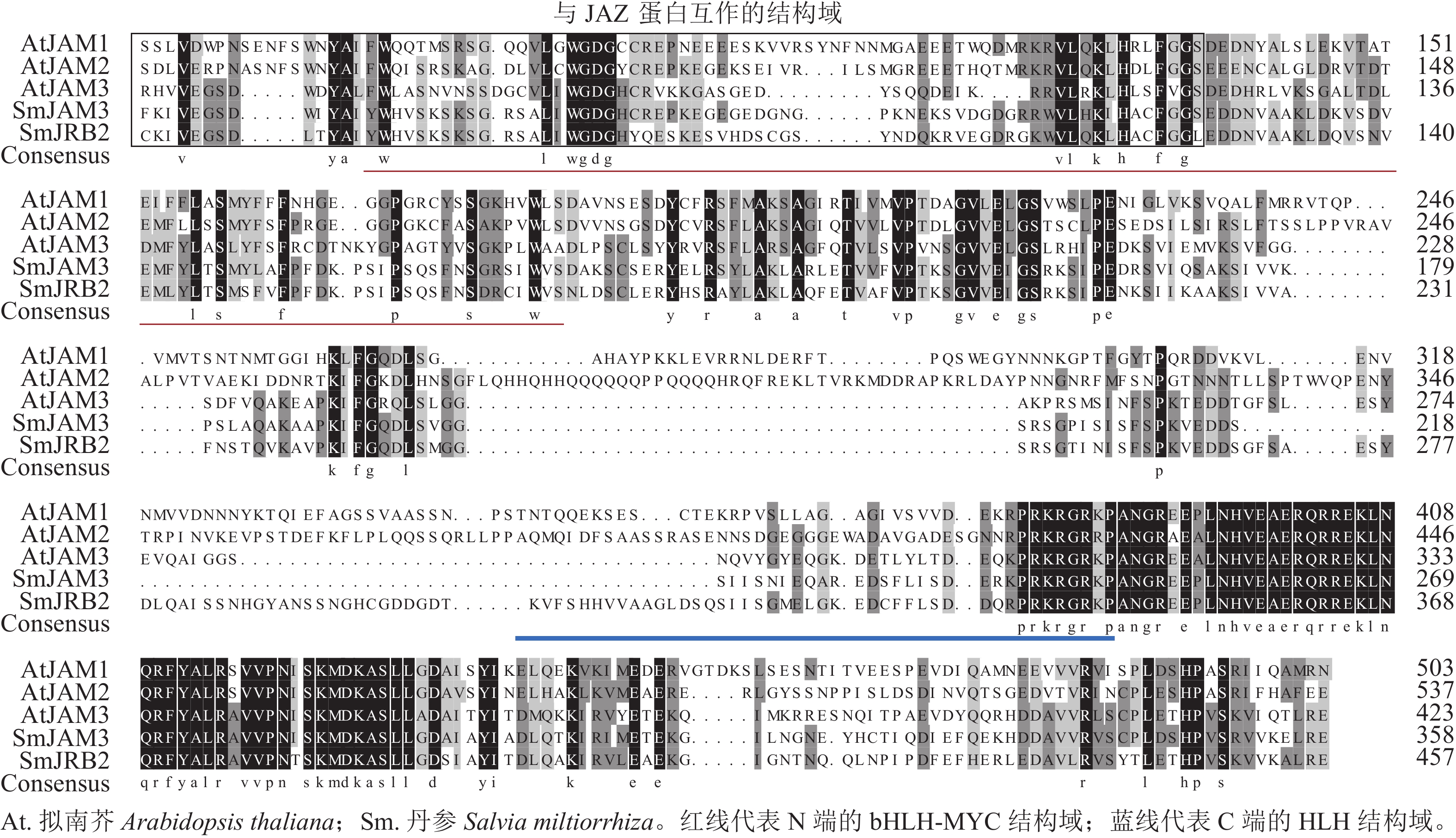
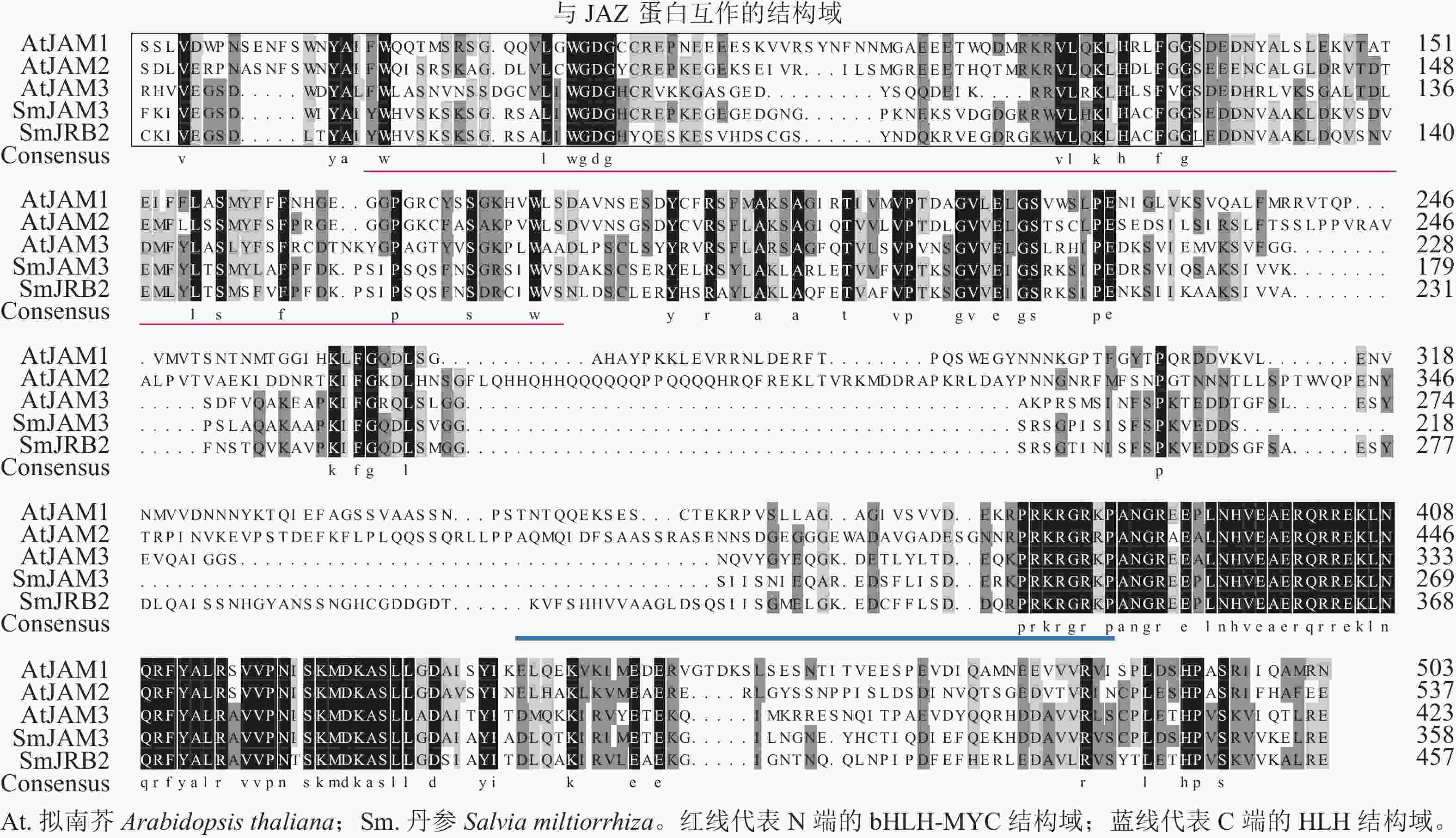
通过PCR扩增获得SmJRB2基因的编码序列,全长为1 506 bp,共编码501个氨基酸,包含71个酸性氨基酸和58个碱性氨基酸;蛋白质分子量为55.57 kD,理论等电点为5.57。对拟南芥Arabidopsis thaliana的AtJAM与 SmJRB2进行蛋白同源比对分析发现:SmJRB2基因蛋白与拟南芥AtJAM3蛋白序列的同源性最高。SmJRB2基因含有2个bHLH-MYC亚家族的保守结构域(图1),其中1个位于N端的bHLH-MYC结构域(114~228个氨基酸区段);另外1个位于C端的HLH结构域(354~405个氨基酸区段),说明SmJRB2极可能属于bHLH转录因子家族的MYC类转录因子。
-
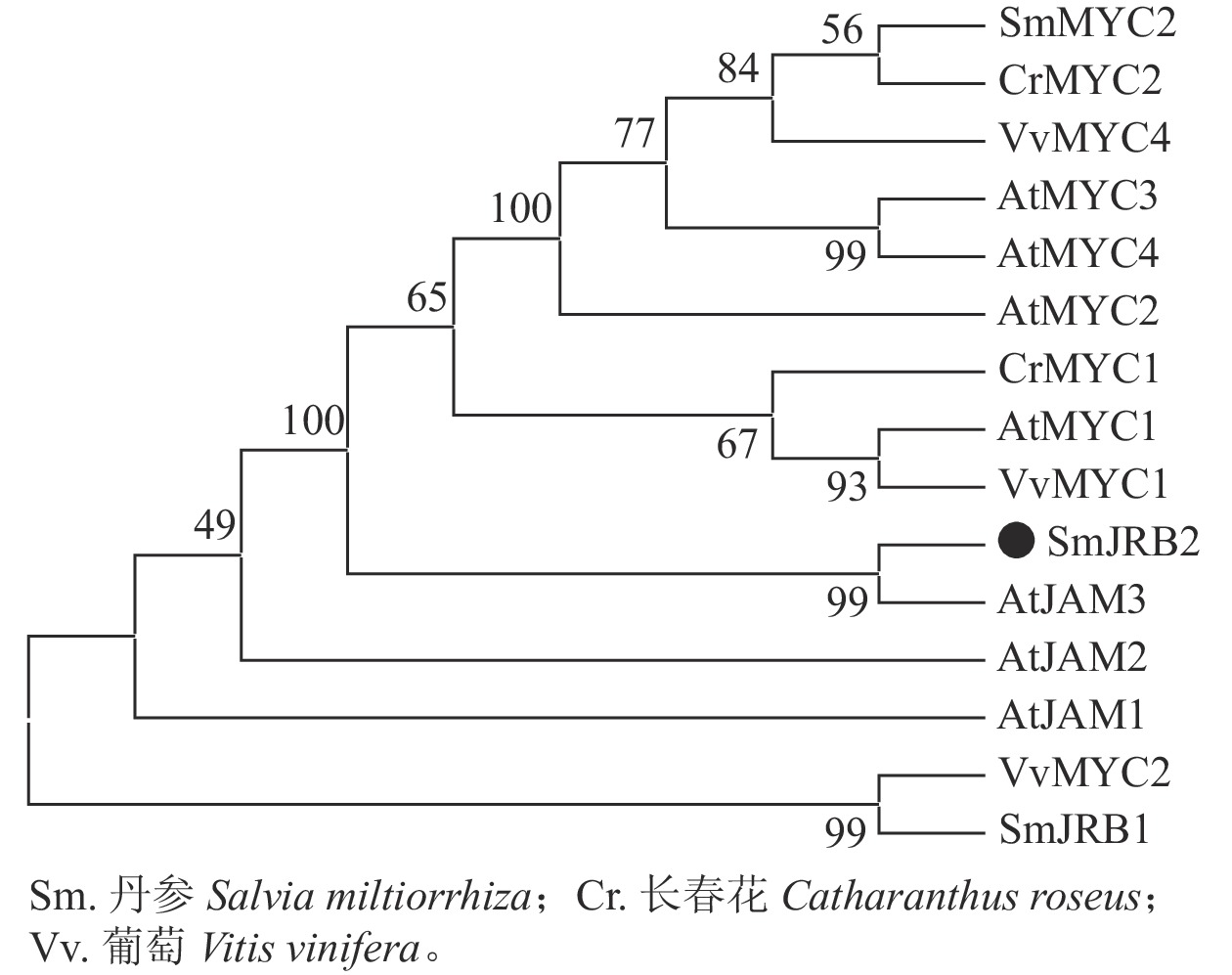
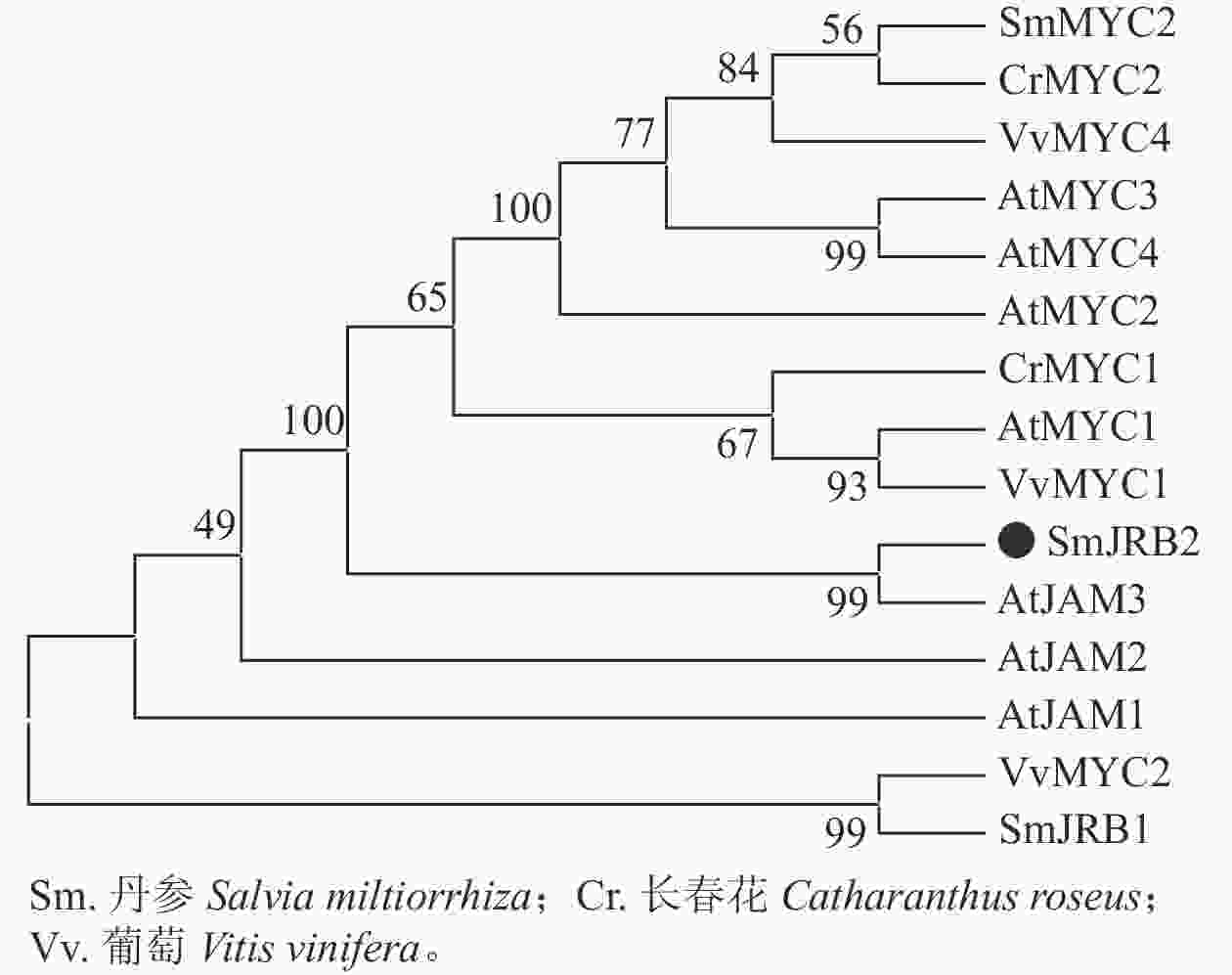
分别以丹参、葡萄Vitis vinifera、长春花Catharanthus roseus及拟南芥AtMYC和AtJAM基因的氨基酸序列为研究对象,利用邻接法构建了SmJRB2蛋白的系统进化树(图2)。结果表明:SmJRB2与拟南芥AtJAM家族成员聚在同一分支,而与MYC类转录因子的亲缘关系相对较远;在拟南芥AtJAM家族成员中,SmJRB2与AtJAM3蛋白具有相对最近的亲缘关系。在拟南芥中,AtJAM3是MYC类转录因子在JA信号通路中基因靶点的竞争子[30−31],说明SmJRB2与AtJAM3具有相似的功能。
-
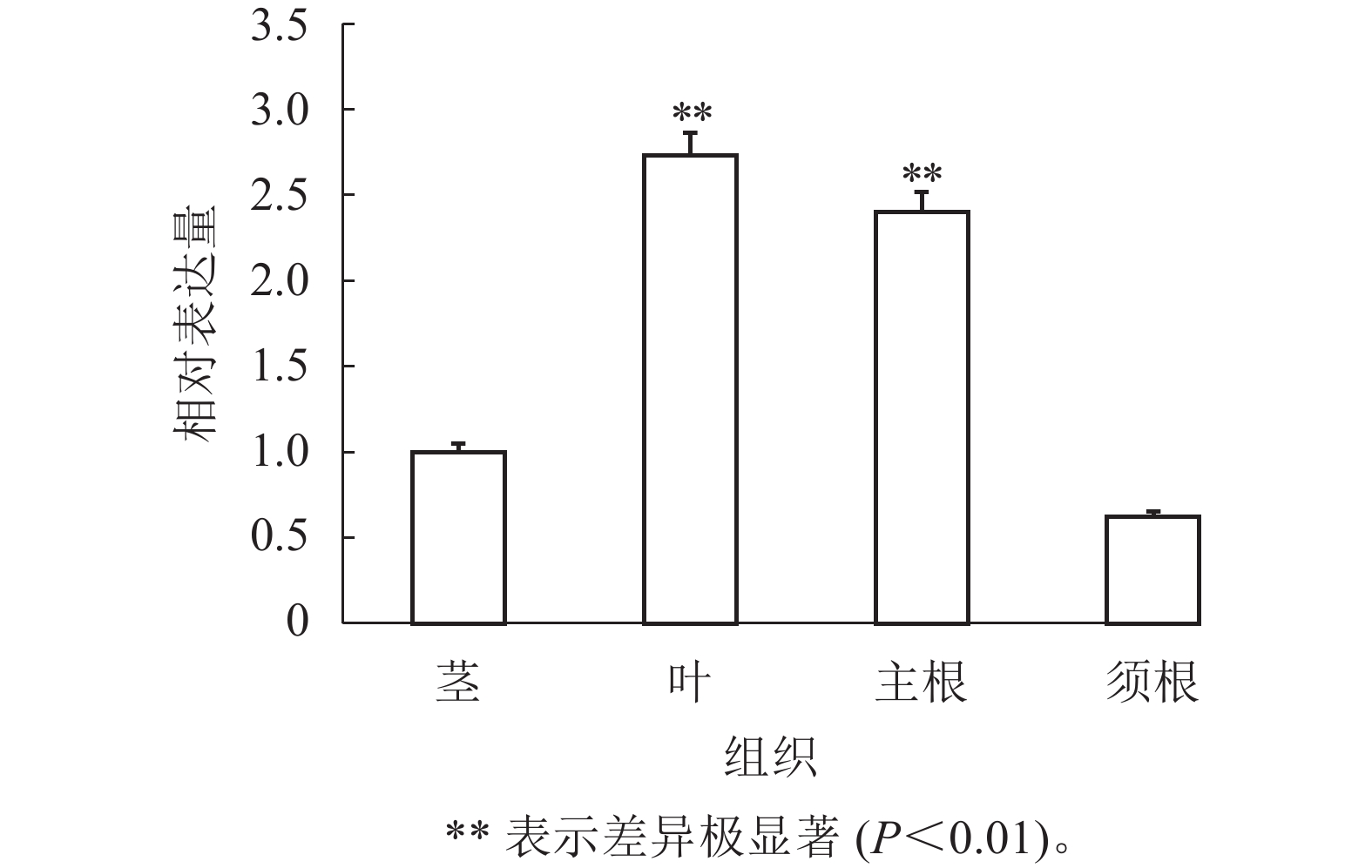
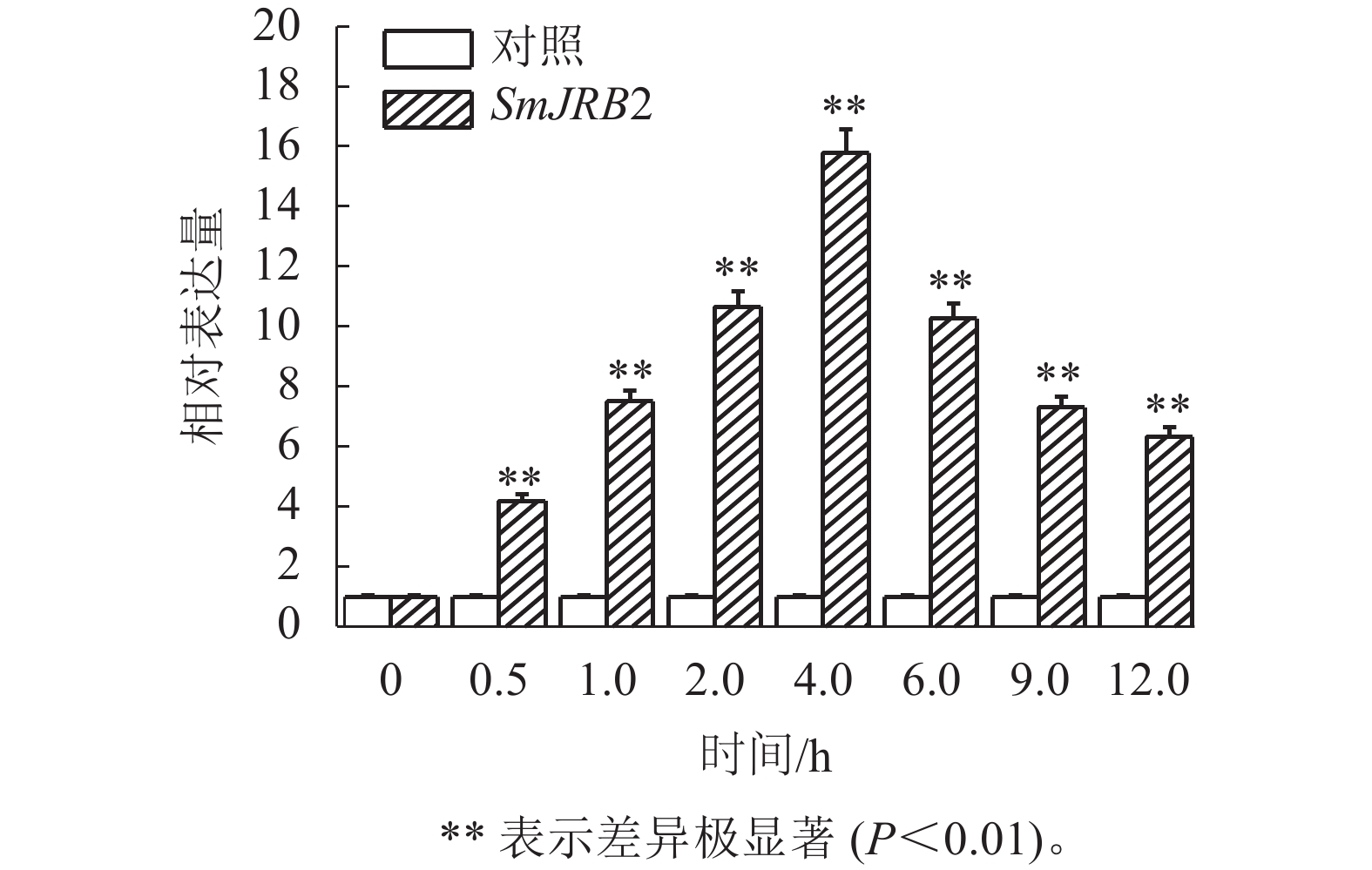
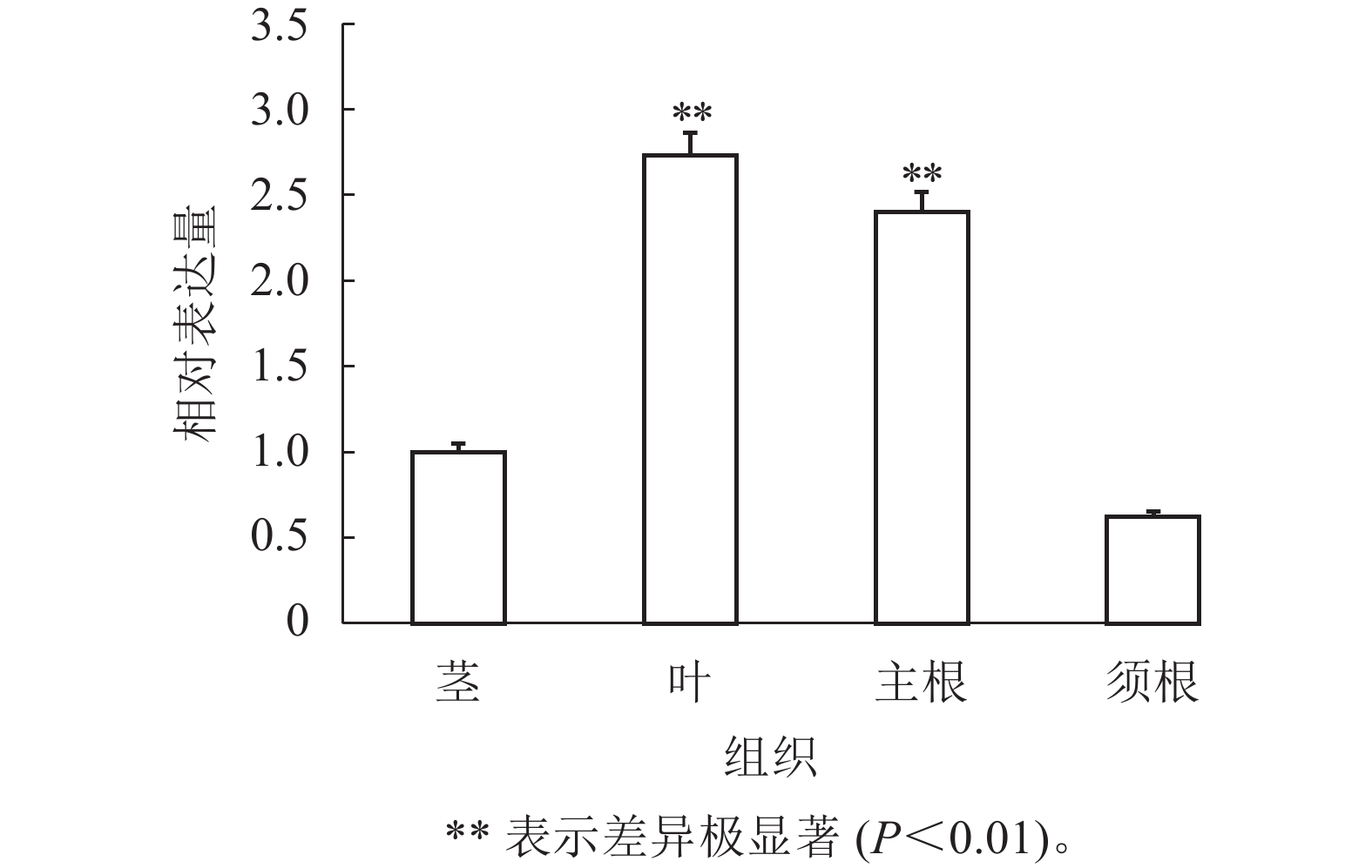
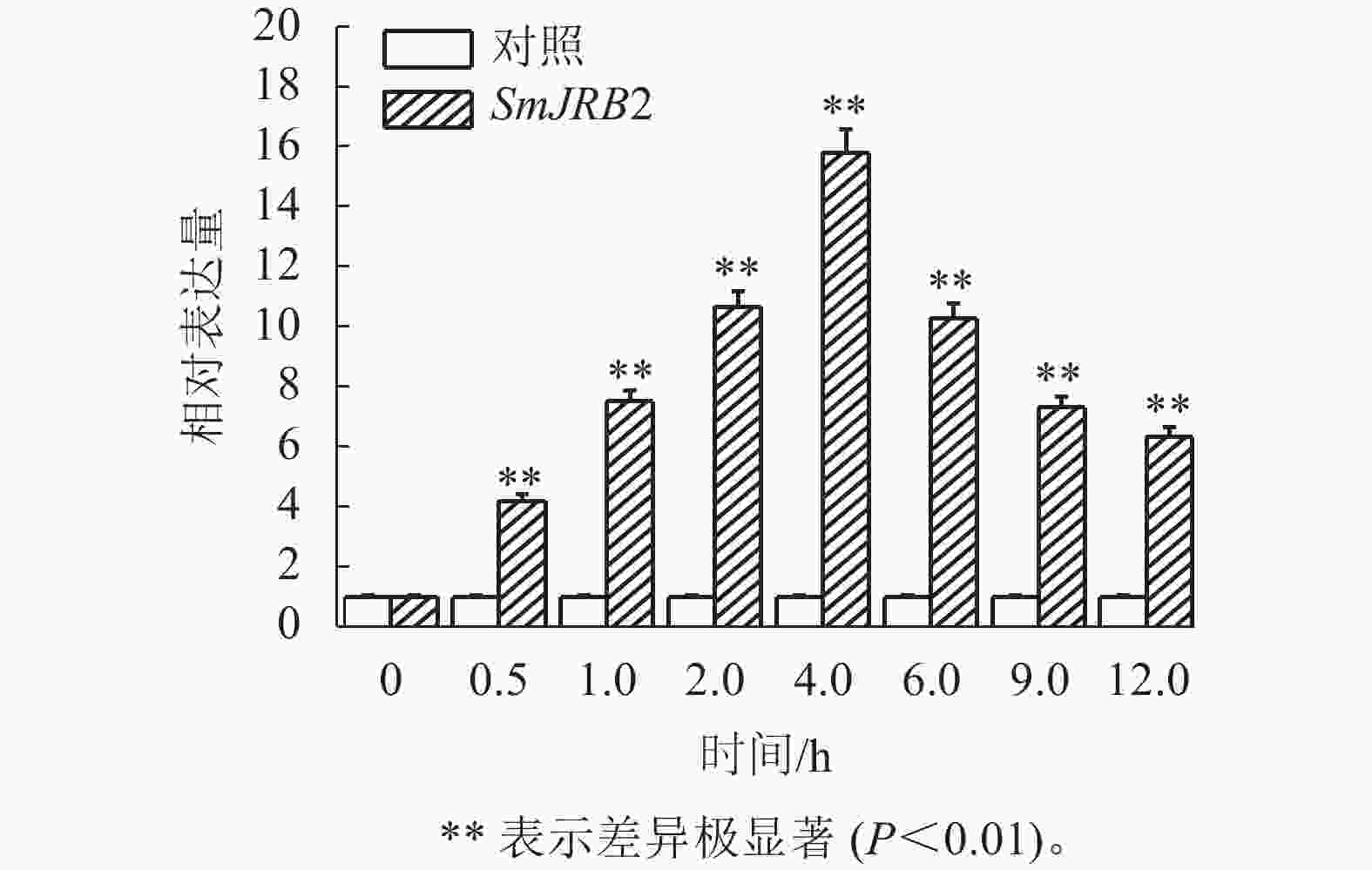
通过RT-qPCR检测SmJRB2基因在丹参各个组织中的表达量。结果表明:SmJRB2基因在叶中表达量最高,在主根中表达量次之,在须根和茎的表达量相对较低(图3)。利用RT-qPCR分析在MeJA诱导下丹参毛状根中SmJRB2基因的表达变化。结果表明:SmJRB2受MeJA诱导0.5 h后,表达量显著上调(P<0.05),而诱导4.0 h时SmJRB2表达量达到最高,上调15倍以上,表明SmJRB2强烈响应MeJA的诱导(图4)。
-
将C58C1空菌株、pHB-SmJRB2-YFP (图5A)+C58C1、pHB-SmJRB2-Anti-YFP (图5B)+C58C1阳性农杆菌活化后,以丹参无菌苗叶片为外植体进行遗传转化。转化后叶片置于1/2 MS+500 mg·L−1头孢噻肟钠(Cef)+100 mg·L−1卡那霉素(Kana)的培养基上培养,每周更换1次培养基;4周后待叶片伤口处出现发根后,再转接到1/2 MS+300 mg·L−1 Cef+100 mg·L−1 Kana的培养基上,每周更换1次培养基;2周后再转到1/2 MS+200 mg·L−1 Cef+100 mg·L−1 Kana的培养基上,每周更换1次培养基;2周后置于无抗生素的1/2 MS培养基上。将2 cm以上的毛状根置于1/2 MS培养基中分株系单独培养,用于阳性检测与扩大培养。
-
以鉴定成功的pHB-SmJRB2-YFP重组子作为阳性对照,以C58C1空菌株诱导的毛状根的DNA为阴性对照,分别检测超表达(SmJRB2-OE)和抑制表达(SmJRB2-Anti)各30个株系中SmJRB2、rolB基因的靶序列。结果表明:SmJRB2过表达阳性株系有8个,阳性率为26.7%;抑制表达阳性株系有9个,阳性率为30.0% (图6)。
-
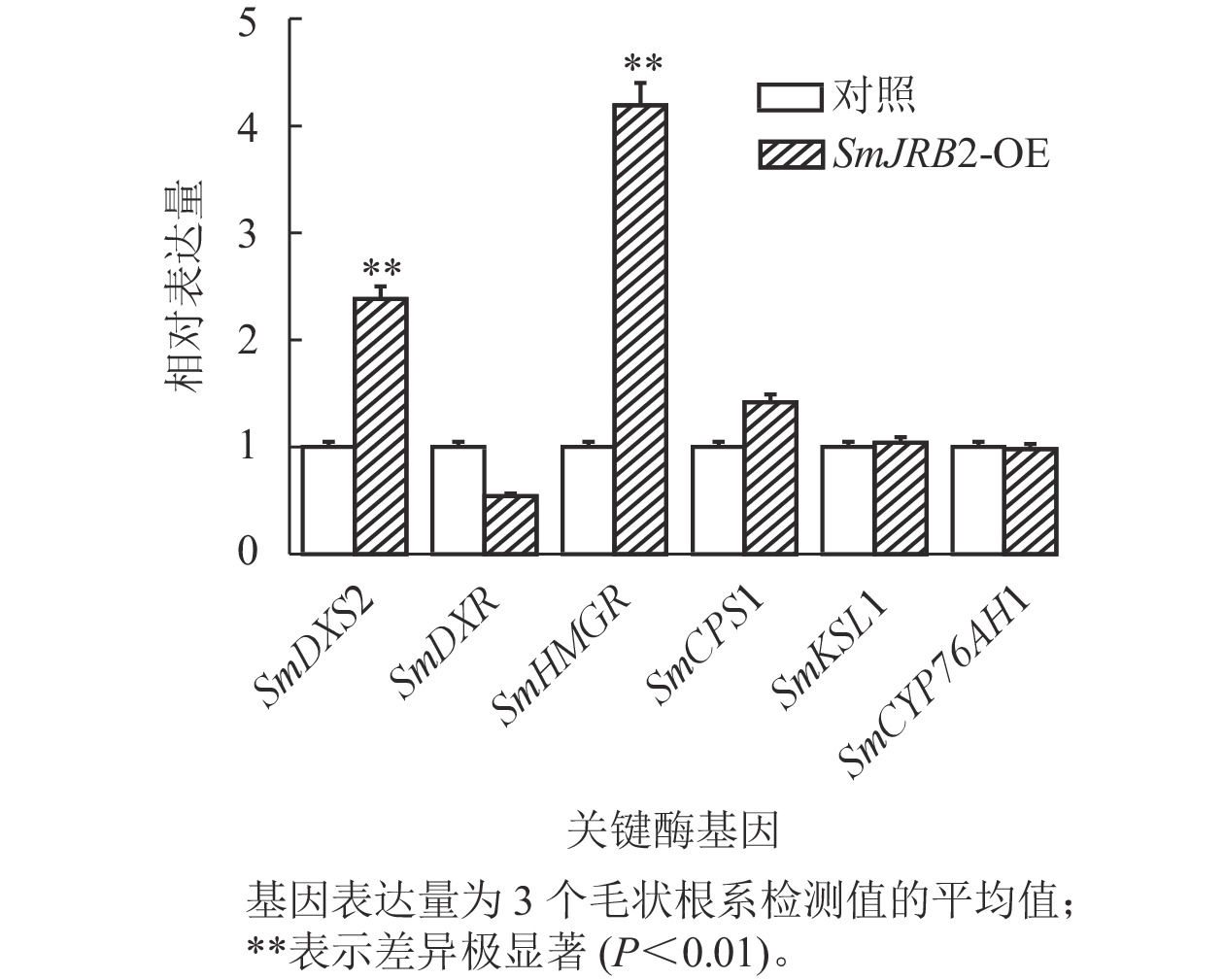
选取3个超表达转基因毛状根系,对SmJRB2过表达毛状根丹参酮代谢途径中关键酶基因的表达量进行RT-qPCR分析。结果显示:丹参酮代谢通路中SmDXS2和SmHMGR基因的表达量显著上调(P<0.05) (图7)。SmJRB2基因可通过上调丹参酮代谢途径的SmDXS2、SmHMGR和SmCPS1等关键酶基因的表达来促进丹参酮的代谢合成。
-
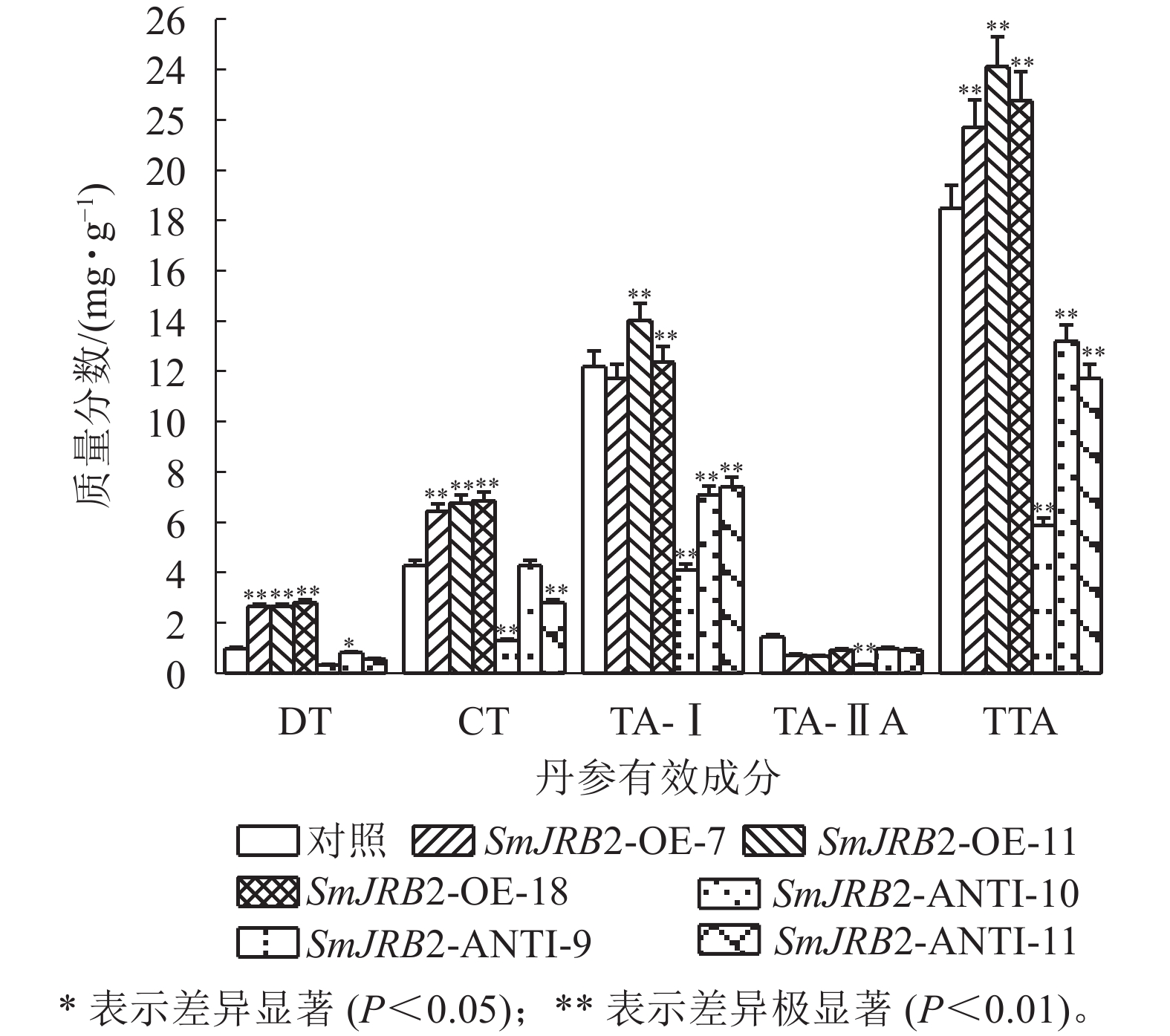
过表达SmJRB2后,3个超表达毛状根系中二氢丹参酮(DT)、隐丹参酮(CT)、丹参酮Ⅰ(TA-Ⅰ)和总丹参酮(TTA)的质量分数均显著提高(P<0.05);3个抑制表达毛状根系中丹参酮DT、CT、TA-Ⅰ和TTA的质量分数均显著下调(P<0.05),TTA质量分数最低,仅为对照的0.31倍;丹参酮ⅡA (TA-ⅡA)质量分数在转基因毛状根中无显著变化(图8) 。结果表明:SmJRB2促进毛状根中丹参酮的积累,是丹参酮代谢的正向调节因子。
-
本研究基于比较转录组测序的结果,采用同源克隆技术克隆了SmJRB2基因的全长编码序列。将SmJRB2基因、丹参转录组数据库获得的SmJRB1基因,以及拟南芥、葡萄、长春花等物种中的MYCs、JAMs转录因子合并构建系统进化树,发现SmJRB2基因与拟南芥的AtJAM3具有较高的同源关系,且含有MYC和JAM保守结构域。基于候选基因的遗传转化实验验证发现,SmJRB2对丹参酮的合成起正向调控作用。YANG等[32]研究发现:MYCs类转录因子作为正向调节子,能显著正调控丹参酮和丹参酚酸的合成。ZHAO等[33]研究发现丹参JAM类转录因子会抑制丹参酚酸的代谢合成。对SmJRB2蛋白的结构域进行分析,结合基因进化特征,推测所克隆的SmJRB2基因属于MYC类转录因子的JAM类亚型,具有其他物种JAM亚型转录调控因子的类似基因功能。
组织表达和诱导表达特征分析表明:SmJRB2转录因子在丹参根、茎、叶中均有表达,且在叶片和主根中的表达量相对最高,在须根和茎中的表达量相对较低。SmJRB2属于MYC类转录因子,叶片中SmJRB2的高表达不仅有利于叶片和茎的生长,还有利于次级代谢产物的生物合成[34−36]。结果显示:SmJRB2能够响应MeJA的诱导,且在诱导后2~4 h内的表达量较高。汪琬宜等[37]和ZHOU等[38−40]研究发现:MeJA是丹参酮和丹参酚酸代谢合成的有效诱导子。因此,推测SmJRB2是MeJA调控丹参药效物质代谢合成的信号响应因子,可为MeJA调控丹参药效物质代谢合成分子机制的阐释提供理论参考。
-
本研究克隆了SmJRB2转录因子,基于农杆菌介导的遗传转化技术分析了该基因的组织表达,发现SmJRB2转录因子在植物的根、茎和叶中均有表达,且在叶中的表达量最高。同时,SmJRB2能响应MeJA的诱导且显著上调。初步阐明了SmJRB2转录因子对丹参次生代谢产物合成的影响,得出SmJRB2是丹参酮代谢合成的正向调控因子。本研究进一步完善了SmJRB2转录因子的调控理论体系,并为丹参优质新品种选育提供了分子靶标和理论基础。
Cloning and functional identification of SmJRB2 gene in Salvia miltiorrhiza
doi: 10.11833/j.issn.2095-0756.20230614
- Received Date: 2023-12-21
- Accepted Date: 2024-03-26
- Rev Recd Date: 2024-03-14
- Available Online: 2024-07-12
- Publish Date: 2024-07-12
-
Key words:
- Salvia miltiorrhiza /
- SmJRB2 /
- cloning /
- expression profile /
- functional identification
Abstract:
| Citation: | JIN Xin, LI Shen, ZHENG Zizhen, et al. Cloning and functional identification of SmJRB2 gene in Salvia miltiorrhiza[J]. Journal of Zhejiang A&F University, 2024, 41(4): 706-714. DOI: 10.11833/j.issn.2095-0756.20230614 |









 DownLoad:
DownLoad: