-
陆地棉Gossypium hirsutum是主要的经济作物之一,在经济发展过程中占有重要地位。而今因优质耕地面积减少导致的粮棉争地问题日益突出,中国陆地棉产区又整体呈现西北内陆棉区面积不断扩大,黄河、长江棉区面积持续减少的趋势[1]。在西北内陆地区栽培早熟棉能充分发挥其可晚播的特点,减少因早春干燥、降温,以及晚霜等原因造成的育苗病虫害,降低杀虫剂施用量[2],增加霜前开花率并改善陆地棉品质[3]。因此,筛选早熟棉对提高耕地利用效率具有重要意义[4]。
植物从生理生长转向生殖生长的过程为开花[5],受环境激素影响[6],目前较为广泛的调控开花途径是光周期途径、春化途径、自主途径及年龄途径等[7]。自主及春化途径主要通过开花抑制基因FLC位点进行[8],FLC调控FT和SOC1抑制开花[9],受光周期途径正向调控[10],可被FLD等通路抑制[11]。光周期靠CO/FT表达改变模式[12]。CO是光周期的核心基因[13],其蛋白有2个锌指结构域正向调控FT[14−15],N端蛋白控制光稳定,C端CCT区域用于核定位[16]。对拟南芥Arabidopsis thaliana研究表明:CCA1/LHY在TOC1上游调控光形态建成抑制其节律[17−18]。激活CCA1/LHY和TOC1翻译组蛋白可调控昼夜节律[19]。RVE8/LCL5也可通过结合TOC1启动子调节昼夜节律[20],节律核心基因限制TOC1的降解[21]。TOC1和CCA1的mRNA转录水平受Hesp调控[22]。
PRR亚家族成员是生物钟重要组分。中心环CCA1和LHY通过结合启动子负调控TOC1(APRR1)[23]。CCA1和LHY是PRR9、PRR7的正调控因子[24],也可能是PRR5的正调控因子。3个PRR基因通过结合启动子负调控CCA1和LHY[25]。PRR5促进TOC1积累使其稳定[26],PRR3和PRR5阻断TOC1与ZTL互作使其稳定[24]。对玉米Zea mays研究表明:PRR家族成员参与包括光响应在内的多种信号传导[27]。对大豆Glycine max研究表明:PRR家族CCT结构缺失与无意义突变影响开花时间[28]。对大白菜Brassica pekinensis[29]、大豆突变体[30]研究也表明:TOC1可控制早花,参与非生物胁迫应答[31]。
陆地棉中开花相关基因大多数属于光周期和生物钟相关途径[32]。前人对陆地棉中光周期通路CO[33]、FT[34],赤霉素途径FPF1[35]、SPL3[36]和部分MADS-box[37−38]家族基因进行研究,说明研究开花通路相关基因具有重要意义。结合生物信息学分析的基因功能研究有助于更好地理解基因功能[39−40]。本研究将从陆地棉群体高密度遗传图谱[41]及数量性状基因座(QTL)定位[42]中发掘陆地棉中拟南芥TOC1(APRR1)的同源基因GhPRR9进行家族分析和功能验证,预测GhPRR9的结构及可能行使的功能,并对GhPRR9功能加以验证,为培育早熟棉提供一定的理论参考。
-
陆地棉全基因组数据下载于Cottongen[43],拟南芥全基因组数据下载于TAIR[44],水稻Oryza sativa、草棉Gherbaceum、可可Theobroma caca、玉米、大豆、毛果杨Populus trichocarpa基因组数据下载于Phytozome[45]。
根据拟南芥PRR亚家族的定义,在Pfam上获得CCT (PF06203)和REC (PF00072)结构域隐马模型,用HMMER扫描整个陆地棉基因组取交集,利用在线工具[46]鉴别所筛选出的基因是否同时包含CCT和REC结构域,最终得到GhPRR家族基因成员。使用ExPASY网站[47]分析工具和WoLF对家族成员进行蛋白理化性质分析。
用MEGA[48]对8个物种的PRR亚家族蛋白进行多序列比对,邻接法JJT模型构建系统进化树,校验重复100次。用DNAMAN进行保守序列比对和绘制。
利用TBtool[49]软件制作染色体定位图和domain结构;使用MEME[50]网站分析家族成员所含Motif并进行可视化。使用Plant Care[51]分析GhPRR亚基因家族上游2 000 bp顺式启动子元件,使用TBtools进行可视化。在美国国家生物技术信息中心(NCBI)数据库中下载陆地棉相关的表达数据(序列号:PRJNA490626,编号:490626),用TBtools绘制热图。
-
提取陆地棉标准系‘TM-1’花蕾RNA,并用试剂盒(CAT#037A)反转录得到底物。使用Primer 5设计引物并扩增目标片段。使用TaKaRa纯化试剂盒(9761)纯化片段,pMD18-T Vector Cloning Kit (CAT# 6011)连接T载。热激法转化DH5α感受态菌株,活化涂板后挑单菌落进行菌液PCR分析,选取合理条带单克隆测序。
以T载为模板克隆片段并连接至过表达载体,热激转化农杆菌Agrobacterium tumefaciens GV3101,筛选阳性单克隆后取带花序的健康拟南芥提前剪下角果。浸入活化农杆菌液侵染1 min,沥干后黑暗1 d正常培养,收集种子为T0代。消毒播种T0代种子至相应抗性培养基上,其中,正常生长幼苗转入正常条件培养。筛选并验证拟南芥的阳性植株,成熟后收取T1代种子,如此培养至T3代。
-
从陆地棉基因组中提取GhPRR9起始密码子上游2 000 bp片段并预测顺式启动子元件。从陆地棉标准系‘TM-1’叶片DNA中分别克隆以起始编码为原点,长500、1 000、1 500和2 000 bp的片段,XcmⅠ酶切链接载体pCXGUS-P,热激法转入大肠埃希菌Escherichia coli,测序无误后将质粒转入农杆菌中侵染拟南芥得到种子。在卡那霉素培养基上播种筛选阳性植株培养至开花,取相关组织染色并观察。
-
将完成转化的表达载体以及绿色荧光蛋白(GFP)空载体通过热激法转入农杆菌菌株GV3101。培养后离心收集菌体重新悬浮,注射幼嫩烟草下表皮。注射后的烟草黑暗培养1 d后恢复正常光照周期。取下表皮制成临时切片,在激光共聚焦显微镜(LSM880)下观察记录影像。
-
相对定量使用2−ΔΔCt法,内参基因为GhHistone3 (陆地棉)和AtUBQ5 (拟南芥)。扩增程序为95 ℃ 30 s,95 ℃ 15 s,60 ℃ 30 s,共40个循环。
陆地棉时空表达分析取样:选取4个品种陆地棉材料的不同器官组织,每个品种20株随机取样,混合研磨。日周期节律分析:取三叶期的陆地棉标准系‘TM-1’植株,在人工气候室中培养1周后,隔4 h取1次顶芽,重复3株混样研磨。
-
用SGN VIGS Tool设计最佳VIGS片段。以测序正确的T载为模板进行扩增,SpeI和AscI酶切位点链接到pCLCrVA载体并转化至农杆菌LBA4404中。pCLCrVA-GhPRR9、pCLCrVA、pCLCrVA-PDS重悬液分别与pCLCrVB的重悬液按体积比1∶1混合均匀。
选取子叶完全展平,第1片真叶尚未完全显形的健康植株进行注射。侵染后的陆地棉设置辅助对照、沉默株、烟草花叶病毒株和空白对照植株,避光培养1 d后转入正常光照培养至开花,记录现蕾开花时间。
-
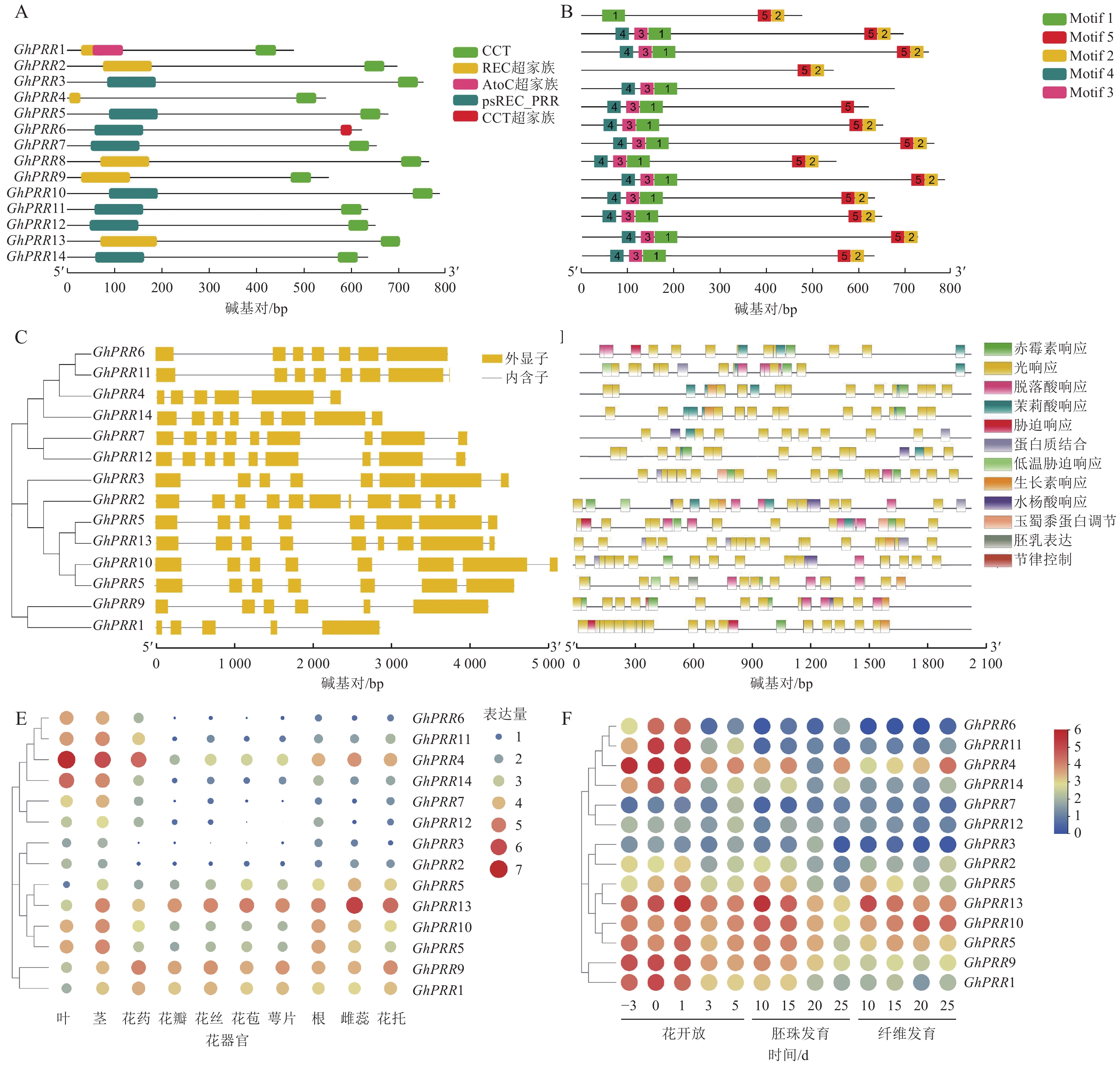
在陆地棉全基因组中共鉴定到14个PRR亚基因家族成员,分别命名为GhPRR1~GhPRR14 (表1)。理化性质分析显示:PRR亚家族成员蛋白有552~775个氨基酸,相对分子量为60.76~85.30 kDa,平均等电点为6.77,酸性蛋白8个,碱性蛋白6个。亚细胞定位结果显示:有11个蛋白定位于细胞核中,2个定位于叶绿体,1个定位于内质网。
蛋白名称 染色体位置 等电点 分子量/kDa 氨基酸/个 亚细胞定位 亲水性 GhPRR1 ChrA03 5.49 53.53 487 细胞核 −0.876 GhPRR2 ChrA05 7.32 76.62 696 细胞核 −0.725 GhPRR3 ChrA05 8.07 81.80 743 叶绿体 −0.738 GhPRR4 ChrA05 8.55 60.76 552 细胞核 −0.834 GhPRR5 ChrA09 6.33 73.66 669 内质网 −0.592 GhPRR6 ChrA11 6.53 68.72 625 细胞核 −0.565 GhPRR7 ChrA11 5.16 73.11 665 细胞核 −0.688 GhPRR8 ChrA11 6.84 82.54 750 细胞核 −0.689 GhPRR9 ChrD03 5.66 61.96 563 细胞核 −0.743 GhPRR10 ChrD09 7.11 85.30 775 叶绿体 −0.677 GhPRR11 ChrD11 7.56 70.20 638 细胞核 −0.692 GhPRR12 ChrD11 5.62 72.77 661 细胞核 −0.622 GhPRR13 ChrD11 7.91 76.08 691 细胞核 −0.680 GhPRR14 ChrD12 6.67 70.92 645 细胞核 −0.742 Table 1. Physicochemical properties of protein in GhPRR subfamily
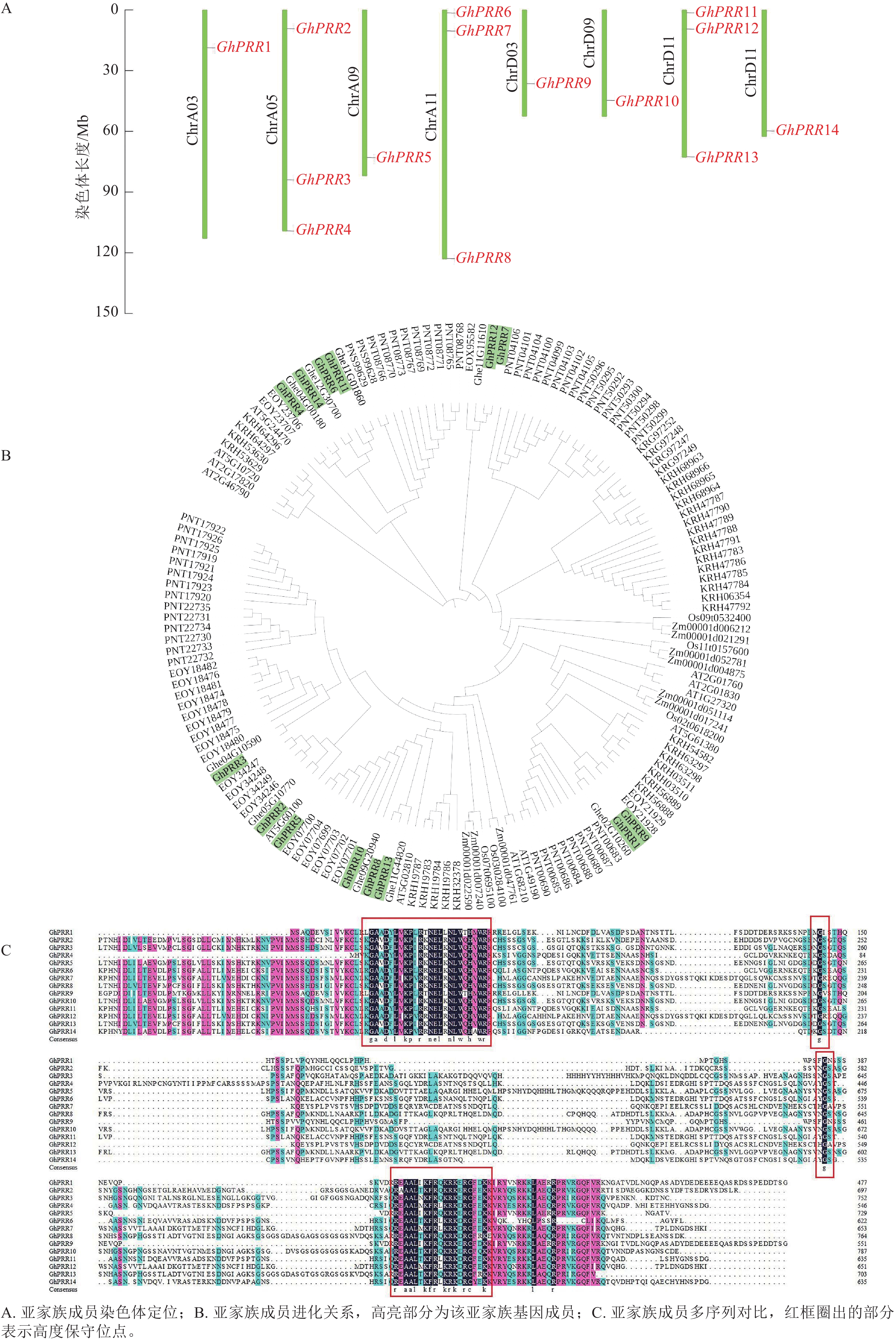
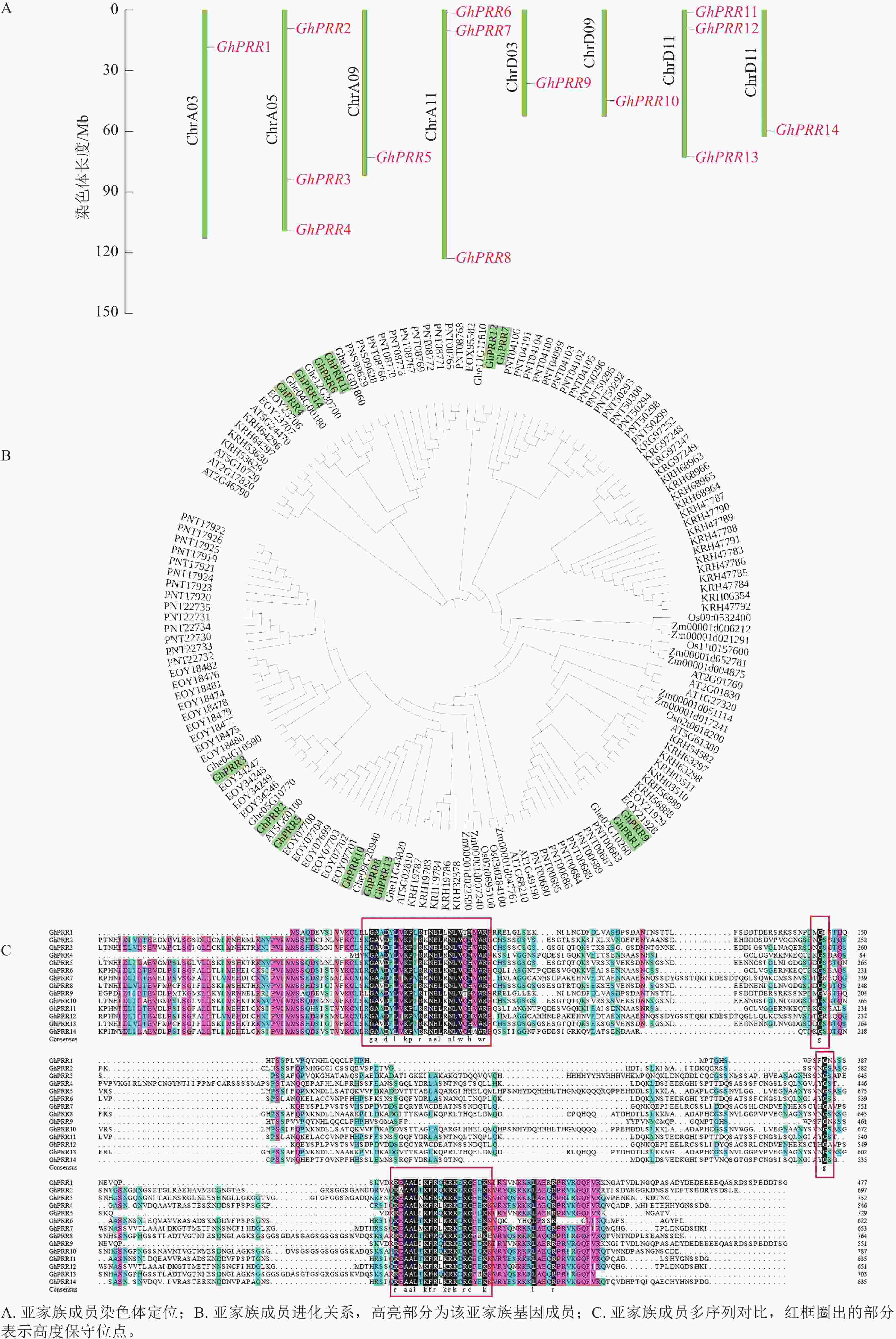
用TBtools绘制出染色体定位图(图1A),可观察到GhPRR家族基因保守分布在染色体两端,14个成员分布在8条染色体上,A亚族8个,D亚族6个,其中Chr A05、Chr A11、Chr D11染色体上分别拥有3个该家族的基因,其余染色体均为1个,表明GhPRR亚家族在陆地棉AD亚基因组上呈现不完全均匀分布。
-
从水稻、拟南芥、玉米、草棉、可可、大豆、毛果杨中分别鉴定出5、6、9、9、24、35、49个PRR亚家族成员,与陆地棉GhPRR亚家族成员蛋白构建系统进化树(图1B)。聚类结果显示:陆地棉GhPRR亚家族进化关系最接近的物种是草棉和可可。不同物种中该基因家族成员的数量差异较为明显,也体现出PRR家族成员在不同物种中的多样性。
多重序列比对(图1C)显示:陆地棉GhPRR亚家族蛋白共有4处位点保守性较强,其中有2个高度保守的g位点,说明该家族拥有2段特征结构域(REC与CCT)。
-
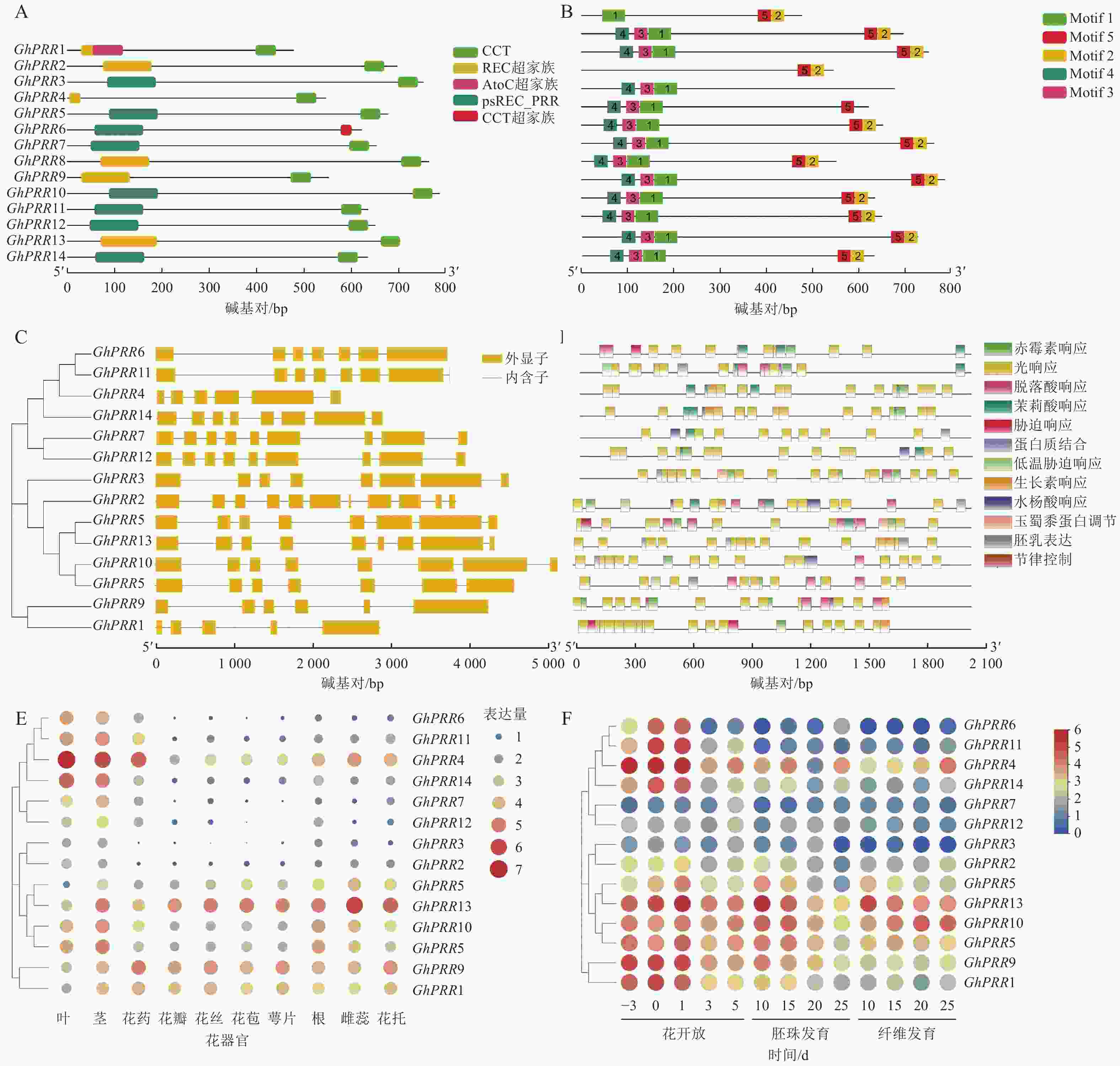
蛋白结构分析显示:14个蛋白均含有CCT结构域(图2A)。GhPRR6、GhPRR1、GhPRR2、GhPRR4、GhPRR8和GhPRR13含有REC超家族结构域(cl19078),其余成员含有psREC_PRR结构域(cd17852,属cl19078超家族),亚家族成员有一定的保守性,REC结构域主要功能为核酸识别,CCT结构域主要标志转录因子,以上2个结构的保守性显示了该家族成员的功能。
MEME分析共得到5个保守基序(图2B)。除GhPRR5外其余成员均含有Motif 2和Motif 5。GhPRR1仅有Motif 1,没有Motif 3和Motif 4,GhPRR4没有Motif 1、Motif 3和Motif 4,其余成员都拥有Motif 1、Motif 3和Motif 4。
基因结构(图2C)显示:该亚家族成员GhPRR1外显子最少(5个),GhPRR2最多(11个),其中,5个成员有8个外显子,3个成员有9个外显子,2个成员有7个外显子,2个成员有6个外显子。最长外显子在3′端较为保守,结构相似度和进化关系基本一致。
-
使用Plant Care对陆地棉GhPRR亚家族成员上游2 000 bp顺式启动子元件进行分析(图2D)发现:主要存在三类顺式元件,一是生长发育响应元件,如光响应元件、生物钟控件;二是激素响应元件,如赤霉素、脱落酸等响应元件;三是非生物胁迫元件,如逆境、盐胁迫等响应元件。其中光响应元件最多(184个),其次为赤霉素响应元件(28个)。说明该亚家族成员主要参与光响应和赤霉素通路。
-
利用公开的转录组数据对陆地棉GhPRR亚家族成员进行组织表达分析(图2E)发现:不同成员组织表达水平差异较大。其中茎叶和花药中表达量最高的是GhPRR4,最少的分别是GhPRR8、GhPRR3和GhPRR2。GhPRR13和GhPRR9在花丝、花苞、花萼中表达量较高,GhPRR2和GhPRR6最少。根中GhPRR13表达量最多,GhPRR7最少,雌蕊花托中GhPRR13表达量最高,GhPRR12和GhPRR2最少。GhPRR4、GhPRR11和GhPRR6可能主要作用于维管组织,GhPRR13和GhPRR9可能作用于花器官组织。
时间表达模式分析(图2F)显示:除GhPRR7、GhPRR12、GhPRR2、GhPRR3和GhPRR10外,GhPRR亚家族其他成员表达量均呈现开花前3 d至开花后1 d逐渐增加,开花后3~5 d逐渐降低的趋势,说明其可能集中在开花前和开花时发挥作用。GhPRR7主要作用在开花后5 d及之后,GhPRR12和GhPRR2可能较少参与开花过程。胚珠中GhPRR13表达量在第10天达到顶峰,说明它在胚珠发育前期可能发挥着一定的作用,GhPRR10、GhPRR5、GhPRR9、GhPRR18、GhPRR3、GhPRR4和GhPRR14也呈现相似的趋势,说明这些基因可能拥有类似的作用模式。纤维发育期间,GhPRR13、GhPRR10和GhPRR4表达量较高,说明这些基因可能参与纤维发育调控。时间模式上,GhPRR13在10~25 d纤维中表达量持续下降,而GhPRR10和GhPRR4则表现出持续上升的趋势,可知GhPRR10和GhPRR4可能参与纤维发育的后期调控,而GhPRR13则参与早期的纤维发育调控。丰富的时空表达说明陆地棉GhPRR家族成员广泛参与到开花前后、胚珠和纤维的发育过程中。
-
使用Plant Care在线工具对该基因上游2 000 bp进行启动子顺式元件分析(表2),发现拟南芥AtTOC1的同源基因GhPRR9上存在着大量的光响应元件,说明光对该基因的转录有着重要的调控作用。除此之外,在GhPRR9基因的启动子区域还存在茉莉酸等激素响应元件,说明该基因可能参与激素相关通路的调节。
名称 起始位置/bp 所在链 功能 名称 起始位置/bp 所在链 功能 ARE 43 − 厌氧胁迫响应 TATA-box 635 − 核心元件 P-box 1 121 − 赤霉素响应 TATA-box 636 − 核心元件 G-box 167 + 光响应 Sp1 1 057 − 光响应 G-box 1 070 + 光响应 G-Box 1 009 − 光响应 A-box 882 − 顺式调节 ABRE 168 + 脱落酸响应 TCCC-motif 871 + 光响应 TGACG-motif 878 + 茉莉酸响应 CAAT-box 249 + 增强区域 TGACG-motif 1 991 − 茉莉酸响应 CAAT-box 354 + 增强区域 Box Ⅱ 1 007 − 光响应 AE-box 535 − 光响应 Box 4 419 + 光响应 GATA-motif 710 + 光响应 MRE 1 513 − 光响应MYB结合 ATCT-motif 1 343 − 光响应 CGTCA-motif 878 − 茉莉酸响应 TATA-box 634 − 核心元件 Table 2. Cis-acting element prediction of GhPRR9 promoter
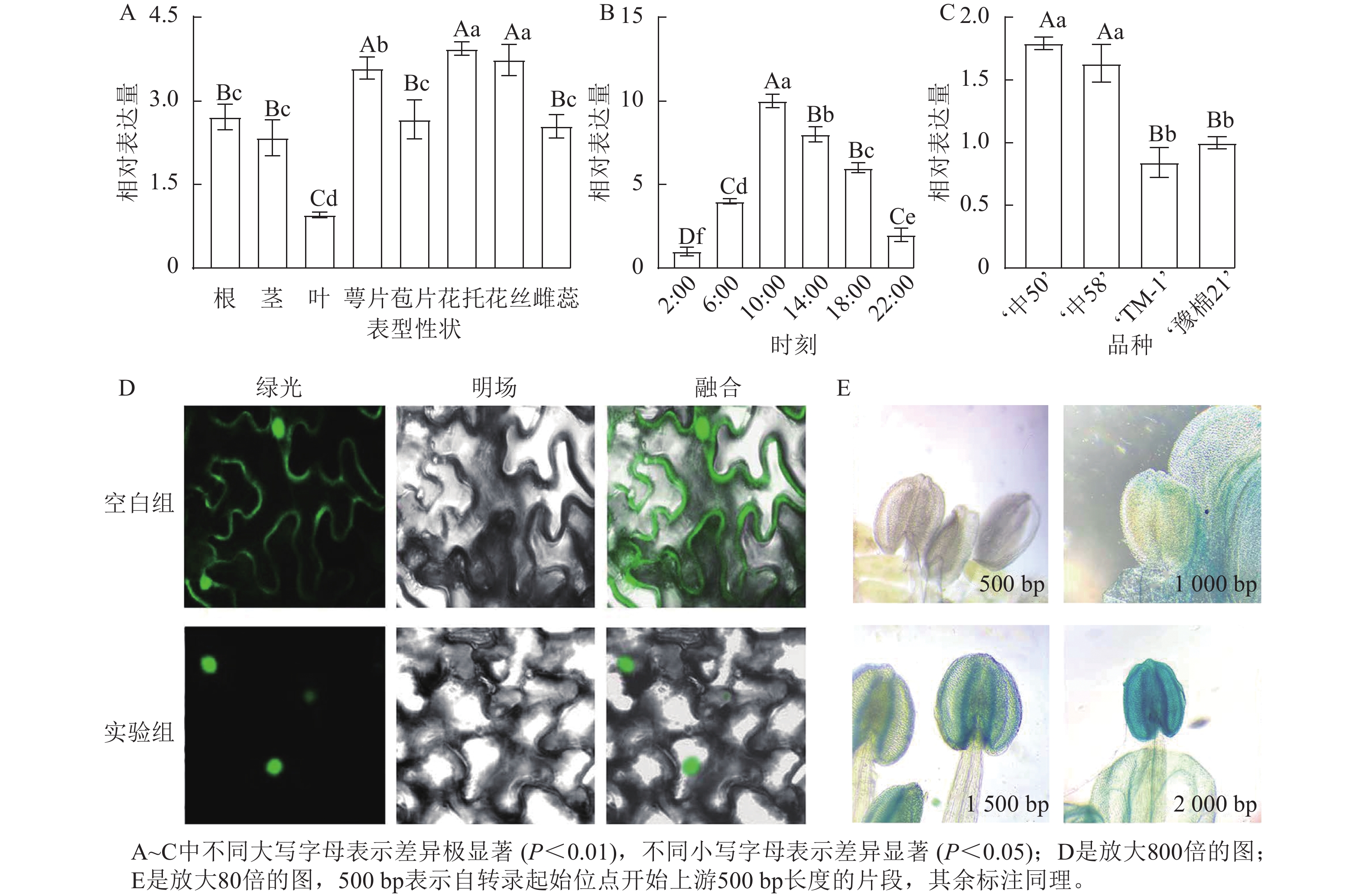
对陆地棉标准系‘TM-1’进行荧光定量分析(图3A)表明:GhPRR9在花丝、萼片、花托中表达量较高,叶片最低,表明GhPRR9可能更多参与陆地棉的生殖生长。
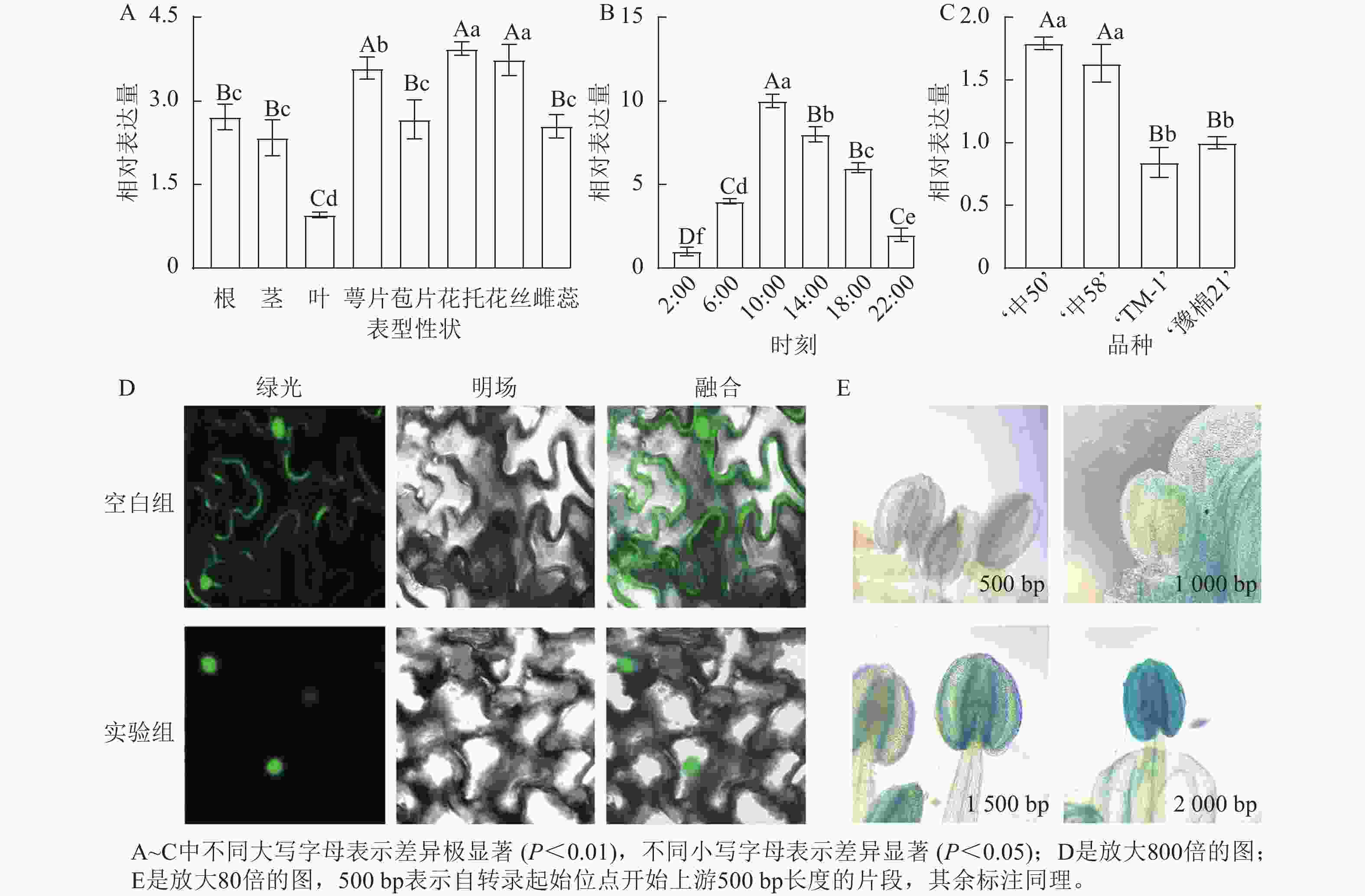
Figure 3. Spatial and temporal expression of GhPRR9 gene, subcellular localization and promoter staining
对在人工光照条件下陆地棉三叶期标准系‘TM-1’顶芽隔4 h取样并进行荧光定量分析(图3B)表明:GhPRR9在光照开始后逐渐积累,并在中午达到顶峰,之后慢慢下降,在光周期内的表达呈现出一定的周期性。
进一步对GhPRR9早熟品种‘中50’‘ZHONG 50’、‘中58’‘ZHONG 58’和晚熟品种‘TM-1’、‘豫棉21号’‘YM21’的表达量分析发现:GhPRR9在早熟品种中表达量显著高于晚熟品种(图3C),说明GhPRR9和早熟性状呈正向相关。
将未转化的GFP质粒和35S::GhPRR9-GFP质粒分别转入农杆菌GV3101,并侵染烟草叶片组织,制作表皮切片置于激光共聚焦显微镜下发现:对照组分布于整个细胞中,而GFP融合蛋白荧光仅分布于细胞核(图3D)。
截取GhPRR9不同长度的启动子与携带GUS报告基因的质粒进行重组,分别转入农杆菌GV3101后通过沾花法侵染拟南芥,获得纯合转基因株系染色观察,结果显示上游500 bp启动子几乎没有表达(图3E),而上游2 000 bp的启动子着色程度最深,说明GhPRR9启动子上游500~2 000 bp内可能存在关键调控元件诱导基因的表达。
-
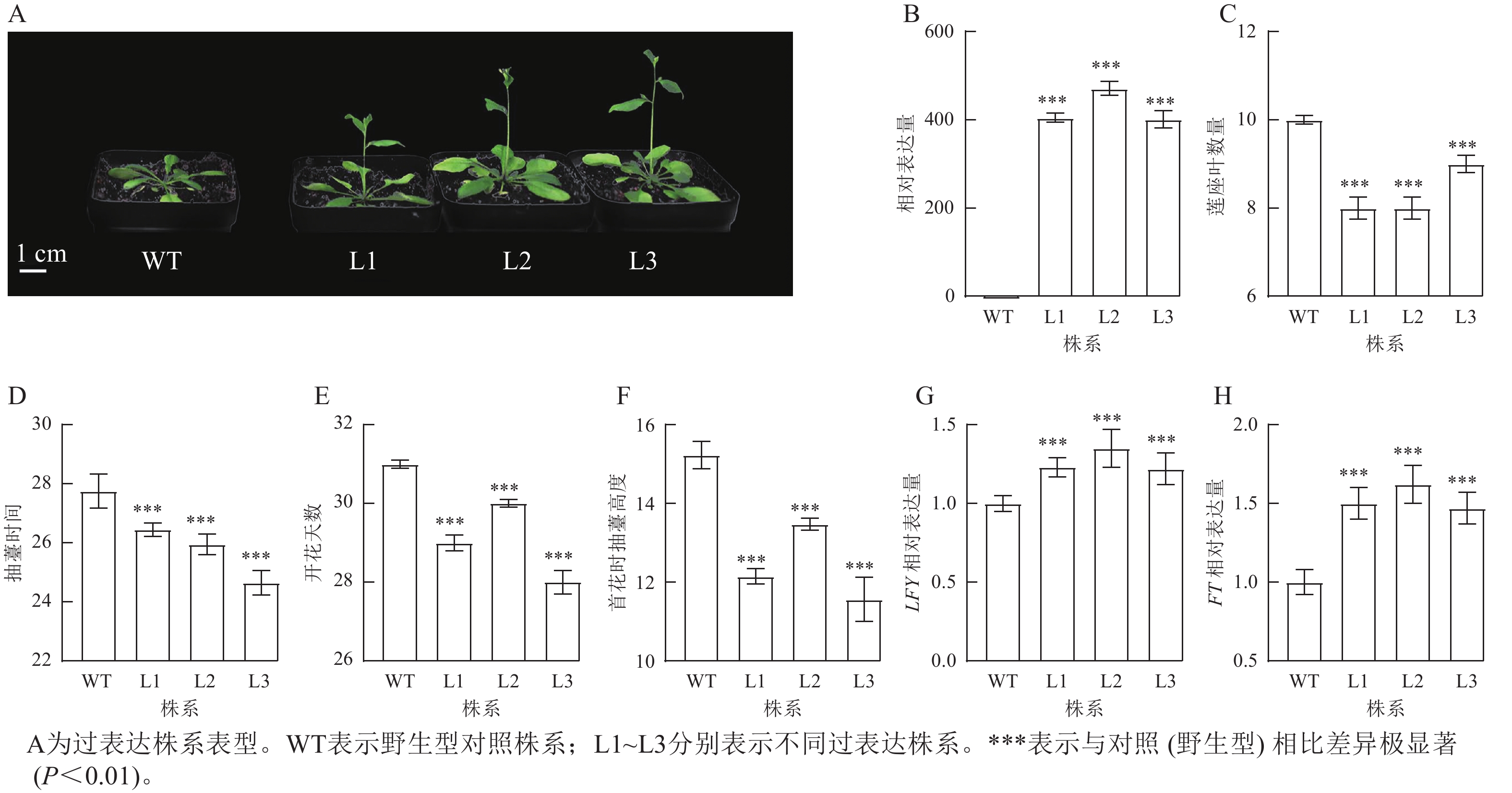
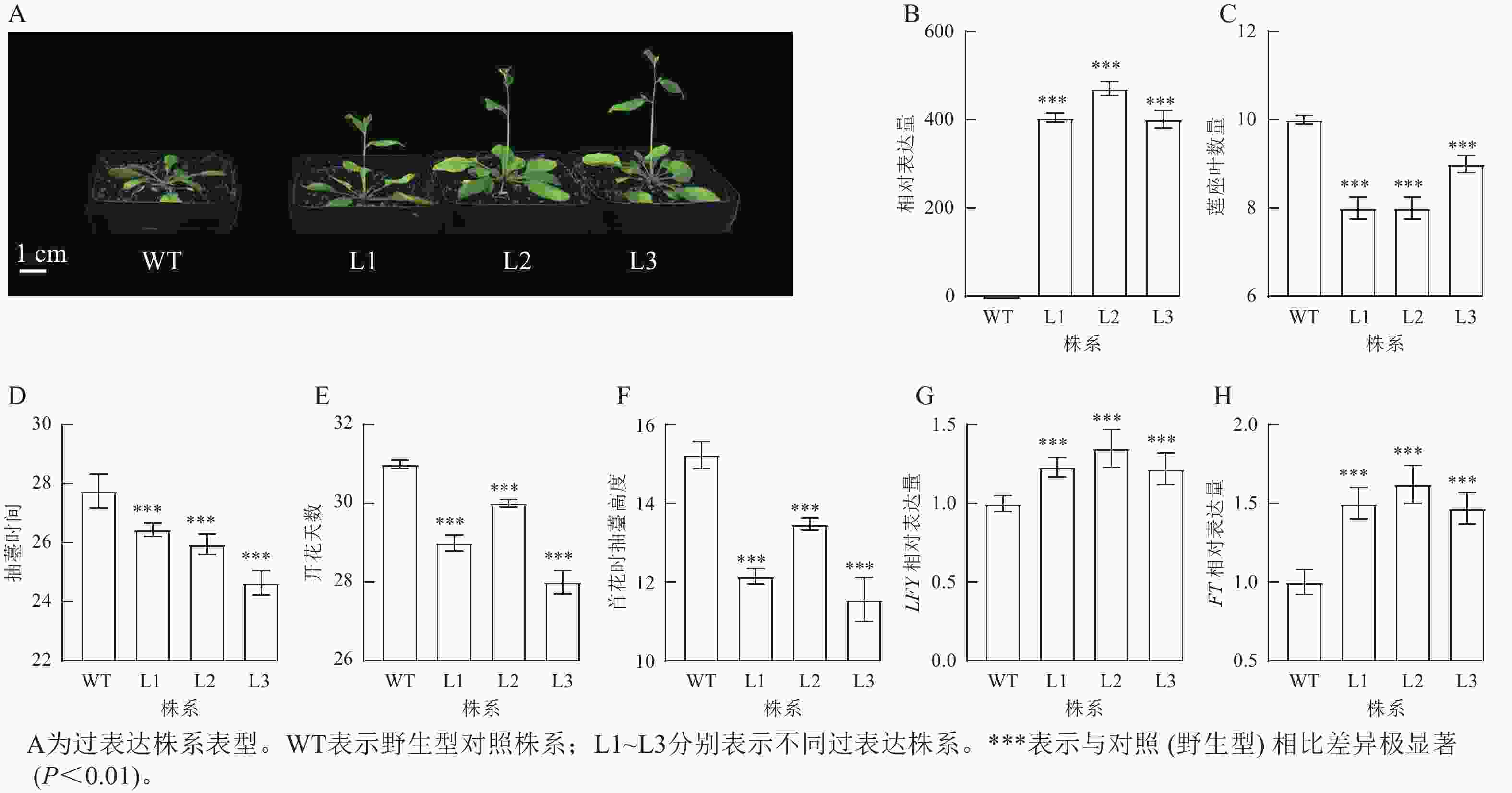
将拟南芥GhPRR9过表达株系培养至抽薹,并观察表型性状(图4A)发现:过表达株系GhPRR9表达量比野生型明显提高(图4B),且转基因过表达株系连座叶数量明显减少(图4C),抽薹时间和开花提前(图4D和图4E),首花抽薹高度极显著矮于野生型(图4F,P<0.01),说明GhPRR9正向调控植物的早花性状。过表达GhPRR9能促进开花关键基因LFY与FT表达(图4G和图4H),表明GhPRR9也可能通过影响关键基因表达调控通路进而影响开花时间。
-
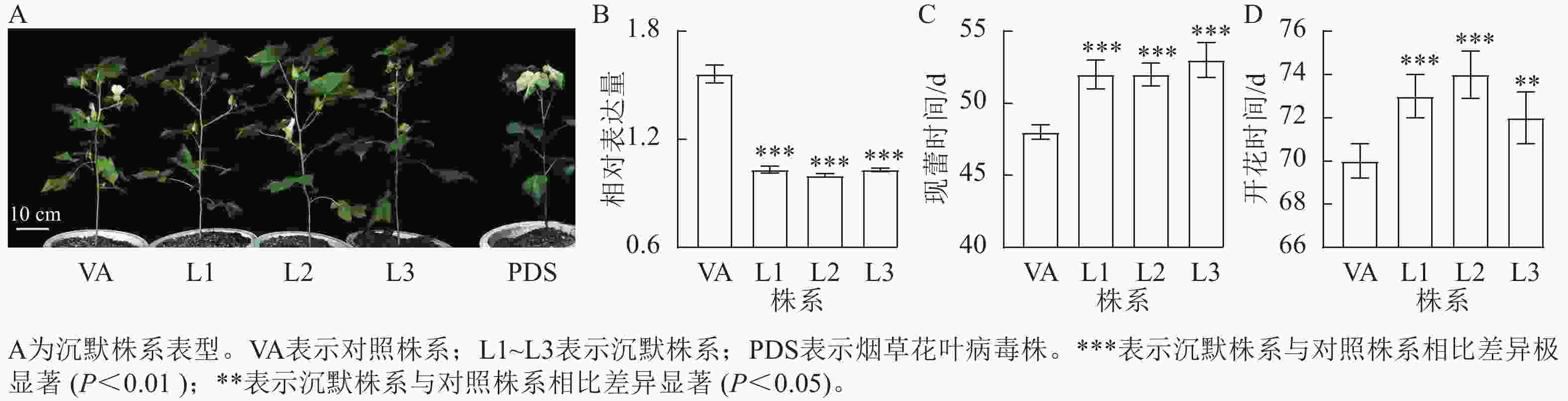
对VIGS沉默株系进行表达量检测发现:沉默株系中,GhPDS株系出现白化表型(图5A),且GhPRR9的表达量极显著降低(图5B,P<0.01),表明GhPRR9基因成功得到了沉默。与对照株系相比,沉默株系现蕾时间延迟约3~4 d,开花时间延迟约2~5 d,表明沉默GhPRR9可推迟开花时间,反向证明其调节陆地棉早花的功能。
-
本研究共鉴定出陆地棉14个GhPRR亚家族成员,成员含有CCT和REC保守结构域,说明其行使转录因子功能。转录组分析显示:大部分成员主要在开花前的茎叶、纤维发育后期和胚珠发育中期发挥作用,表明大部分成员可能存在功能冗余或协同拮抗作用。启动子元件分析显示:陆地棉GhPRR亚家族可能频繁地参与光感效应相关的生理过程,这与在拟南芥的结果中一致,据此可推测其与拟南芥同源基因作用相似。进化分析表明:陆地棉GhPRR亚家族成员基因数量多于拟南芥。前人研究也发现:棉花基因组进化加倍使该家族基因得到了扩增[52]。
对过表达株系研究发现:抽薹日期、开花日期都稍有提前,抽薹高度显著高于同期野生型植株,证明GhPRR9可以使拟南芥花期提前。构建GFP表达载体侵染烟草叶片,表明GhPRR9蛋白定位于细胞核,与生物信息学分析相互印证,进一步确认该基因行使转录因子的功能。对GhPRR9基因1 d内表达水平分析显示:光暗交替条件下基因表达量存在着周期性变化,按照其表达模式推断该基因在光照开始后积累,中午达到顶峰后慢慢下降,这与拟南芥同源基因的表达模式[53]相似,据此推测,陆地棉早花基因GhPRR9可能与拟南芥中的同源基因行使着类似的功能。陆地棉三叶期叶片的基因表达量结果显示:GhPRR9基因在早熟种中表达量高于晚熟种,据此可推断其与早熟性状有正向关联。构建VIGS株系发现:沉默株系高度降低,生育期推迟,反向证明了其促进生育期的功能。启动子分析显示:光和赤霉素可能影响该基因的转录。GUS染色结果显示:启动子上游500~2 000 bp可能存在关键调控元件。有研究显示:陆地棉转录因子与通路主要基因启动子的结合可随温度产生变化[54],且同源转录因子可能存在相互调控的作用[55]。
-
本研究预测了陆地棉GhPRR亚家族的功能和作用模式,找到拟南芥早花基因AtTOC1的陆地棉同源基因GhPRR9,并成功克隆,构建遗传转化株系对其功能进行验证显示:GhPRR9对早花性状存在正向促进作用。
Functional analysis and validation of early flowering gene GhPRR9 in Gossypium hirsutum
doi: 10.11833/j.issn.2095-0756.20240267
- Received Date: 2024-03-31
- Accepted Date: 2024-10-14
- Rev Recd Date: 2024-06-21
- Available Online: 2025-01-20
- Publish Date: 2025-02-20
-
Key words:
- Gossypium hirsutum /
- family analysis /
- GhPRR9 /
- functional verification /
- early blossoming
Abstract:
| Citation: | ZHANG Ya’nan, XU Tingting, XU Haobiao, et al. Functional analysis and validation of early flowering gene GhPRR9 in Gossypium hirsutum[J]. Journal of Zhejiang A&F University, 2025, 42(1): 74−85 doi: 10.11833/j.issn.2095-0756.20240267 |









 DownLoad:
DownLoad: