-
桂花Osmanthus fragrans是中国十大名花之一,深受人们喜爱。前人研究报道桂花花香主要成分为α-紫罗兰酮、β-紫罗兰酮、芳樟醇及其氧化物、罗勒烯等挥发性物质[1],而桂花花色主要成分是类胡萝卜素化合物[2]。桂花的花香和花色是其最主要的观赏性状,与花香和花色相关次生代谢物质的合成与积累直接影响到桂花的观赏品质。植物呼吸作用不仅提供植物所需的能量,一系列的中间产物还为多种次生代谢化合物合成提供原料,在植物体内有机物转变方面起到枢纽作用。已糖激酶(hexokinase,HXK)具有己糖磷酸化功能,经HXK磷酸化的己糖才能进行植物呼吸代谢的糖酵解途径,为植物生长和发育提供能量和中间代谢产物[3-6]。高等植物HXK基因以多基因家族形式存在[7-11]。拟南芥Arabidopsis thaliana的6个HXK家族成员中,AtHXK1~3能磷酸化葡萄糖[10],其中AtHXK1还具有感知和传递己糖信号的功能[12-13]。AtHXK1在强光条件下对植物根、叶和花序的生长和发育有促进作用[13]。此外,拟南芥成花转变[14]和种子萌发[15]、植物花青素积累[16]和脂肪族硫甙生物合成[17]等植物生命活动都与HXK介导的己糖信号转导息息相关。目前,未见桂花HXK基因功能有关的研究报道。本研究基于桂花不同花色品种转录组数据筛选OfHXKs相关Unigene序列,分析不同OfHXKs基因家族成员核苷酸序列及其编码的氨基酸序列,并与其他物种HXK蛋白进行多重比对和进化树分析,同时分析不同桂花品种不同发育阶段花瓣以及不同组织中OfHXKs基因表达特征,为进一步揭示桂花不同OfHXKs基因成员功能奠定理论基础。
-
选择浙江农林大学桂花资源圃生长状况良好的地栽桂花‘堰虹桂’Osmanthus fragrans ‘Yanhong Gui’(YHG)、‘玉玲珑’O. fragrans ‘Yulinglong’(YLL)、‘金球桂’O. fragrans ‘Jinqiu Gui’(JQG)为材料,依据桂花花序不同发育阶段[18],对这3个桂花品种顶壳期(S1)、铃梗期(S2)、初开期(S3)、盛开期(S4)花瓣分别进行取样,取样时间均为10:00。同时还对‘堰虹桂’1年生茎、2年生茎、嫩叶、成熟叶和盛开时花序进行取样,用于不同组织中OfHXKs基因表达分析。用液氮速冻暂存样品,后存于−80 ℃冰箱。
-
RNA提取试剂盒采用RNAprep Pure Plant Kit(天根,北京);反转录试剂盒使用PrimeScript™ RT Master Mix(Perfect Real Time)(Takara,大连);荧光定量试剂盒使用TB Green® Premix Ex Taq™ Ⅱ(Tli RNaseH Plus)(Takara,大连)。实时荧光定量PCR仪为 LightCycler®480 Ⅱ(Roche,德国)。
-
从转录组中筛选得到HXK相关的Unigene序列,首先使用美国国家生物信息中心(NCBI)数据库中的BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi)对得到的基因序列进行对比;利用ORF FINDER (https://www.ncbi.nlm.nih.gov/orffinder/)寻找开放阅读框;用DNAMAN 6.0软件对不同品种相同HXK基因成员进行核苷酸相似性比较,使用‘堰虹桂’的4个HXK基因成员序列开展后续序列分析,包括应用Prot-Param在线软件(http://web.expasy.org/protparam/)预测所编码蛋白的分子量、理论等电点(PI)、不稳定系数、总平均亲水性等;使用YLoc 进行亚细胞定位预测;使用SOPMA分析软件对OfHXKs家族的蛋白序列进行二级结构预测分析,判断目的基因的氨基酸残基是否处于于α-螺旋、β折叠(或其他状态)的二级结构;用SWISS-MODEL (https://swissmodel.expasy.org/)预测OfHXKs的三级结构;应用SignalP 5.0 对信号肽进行预测;利用TMHMM在线软件预测分析桂花蛋白的跨膜结构;使用NetPhos 3.1 Server 对桂花花瓣OfHXKs蛋白质磷酸化位点进行预测;采用MEGAX软件中的邻位相邻法(neighbor-joining,NJ)进行同源聚类,建立系统发育树,并采用Bootstrap法(重复1 000次)评估检测系统进化树。
-
采用RNAprep pure Plant Kit(天根,北京)多糖多酚植物总RNA提取试剂盒按照说明书步骤提取各样品的总RNA。使用紫外分光光度计和质量浓度为1%琼脂糖凝胶电泳检测总RNA浓度和质量。反转录体系为总RNA 1.0 μL,5×PrimeScript RT Master Mix(TaKara,大连)2.0 μL,无RNA酶双蒸水7.0 μL,PCR反应的程序设定为:37 ℃ 15 min,85 ℃ 5 s,4 ℃ 1 min。反转录合成的cDNA储存于−20 ℃备用。
-
根据荧光定量PCR引物设计原则利用Primer Premier 5进行引物设计,以桂花OfACT为内参基因[19],引物序列见表1。荧光定量PCR反应体系为:TB Green Premix Ex Taq Ⅱ(TaKaRa,大连) 5.0 μL,上下游引物(10 μmol·L−1)各0.4 μL,cDNA模板(反转录cDNA稀释20倍)2.0 μL,双蒸水2.2 μL,每个样品设置3个生物学重复。反应程序为两步法:95 ℃预变性30 s,95 ℃变性5 s,60 ℃复性30 s,重复40个循环;然后以95 ℃持续5 s,60 ℃持续1 min,95 ℃持续15 s作为溶解曲线程序。
表 1 OfHXKs基因表达分析所用引物序列
Table 1. Primer sequences of OfHXK genes of O. fragrans
基因名称 正向引物序列(5′→3′) 反向引物序列(5′→3′) OfHXK1 TTCTTCTTCGTCTGGCGTTCTG GTGCATTAACCCGCATATCCAGG OfHXK2 ACCTCCCTAAAAACAAGGAGAGTTG AGTATCCCGTCCCATTTTCTTTAGG OfHXK3 CACTTATTTGGTCACTCAGTTCCCG ACACACGTCTATGACAATCTTCCTC OfHXK4 GCACTCATTGCAGCCTCTCACT CTCACTCTGACAGTGACCGGCGTTA OfACT CCCAAGGCAAACAGAGAAAAAAT ACCCCACTACCAGAATCAAGAA -
根据桂花不同花色品种转录组Unigene序列分析,筛选得到4个HXK基因的同源基因(OfHXK1~OfHXK4),3个品种中均含有这4个HXK基因的同源基因。利用DNAMAN对不同品种OfHXK1~OfHXK4核苷酸序列进行多序列比对,核苷酸序列相似性比较结果见表2。不同品种桂花OfHXK1、OfHXK3和OfHXK4基因核苷酸序列相似性均高于99%,而不同品种桂花OfHXK2基因核苷酸序列相似性为94.42%~98.26%。选择‘堰虹桂’的OfHXK1~OfHXK4基因推测的氨基酸序列进行后续分析。
表 2 桂花OfHXKs核苷酸相似性比较表
Table 2. Comparison of nucleotide sequences of OfHXKs in O. fragrans
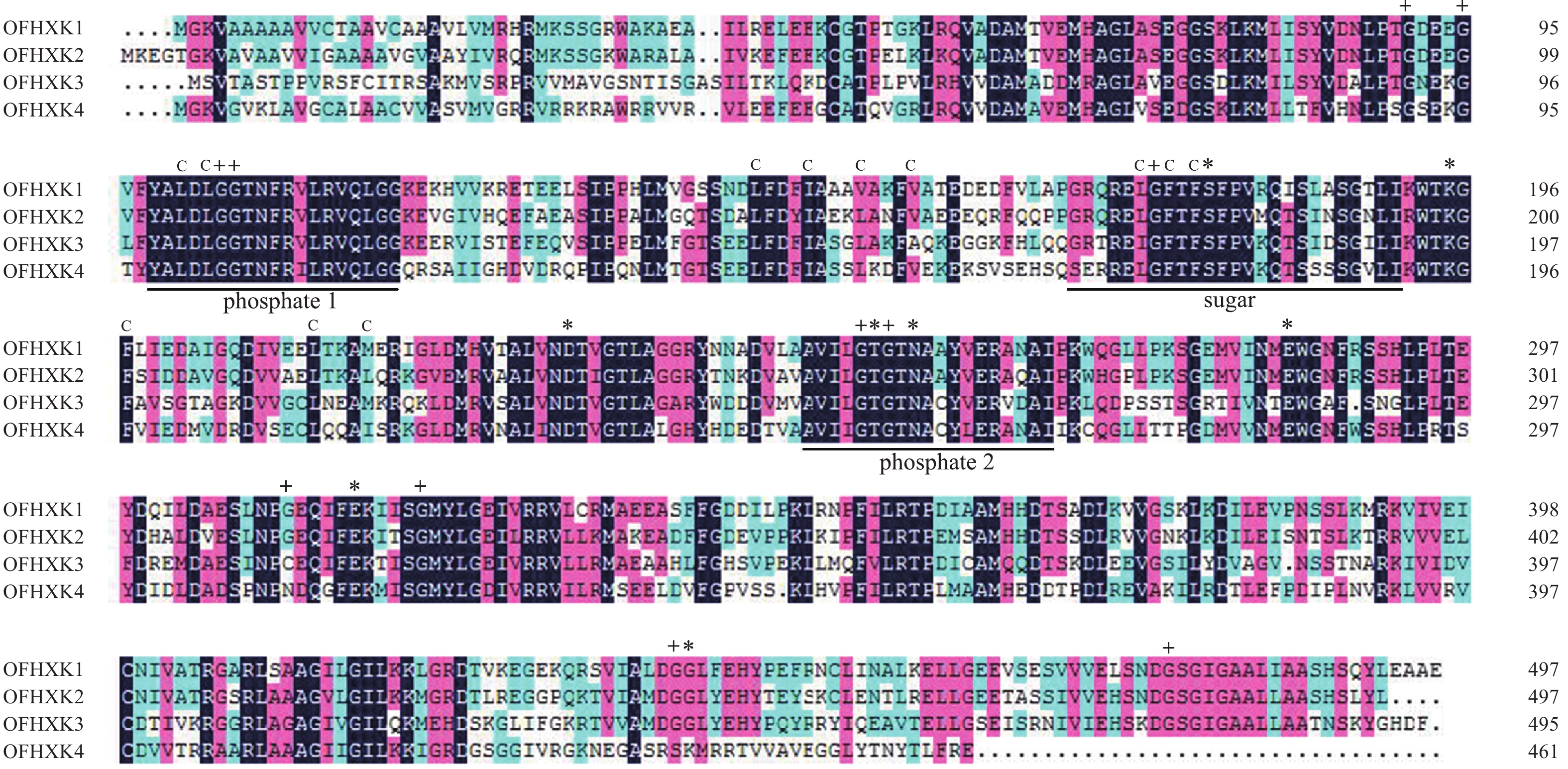
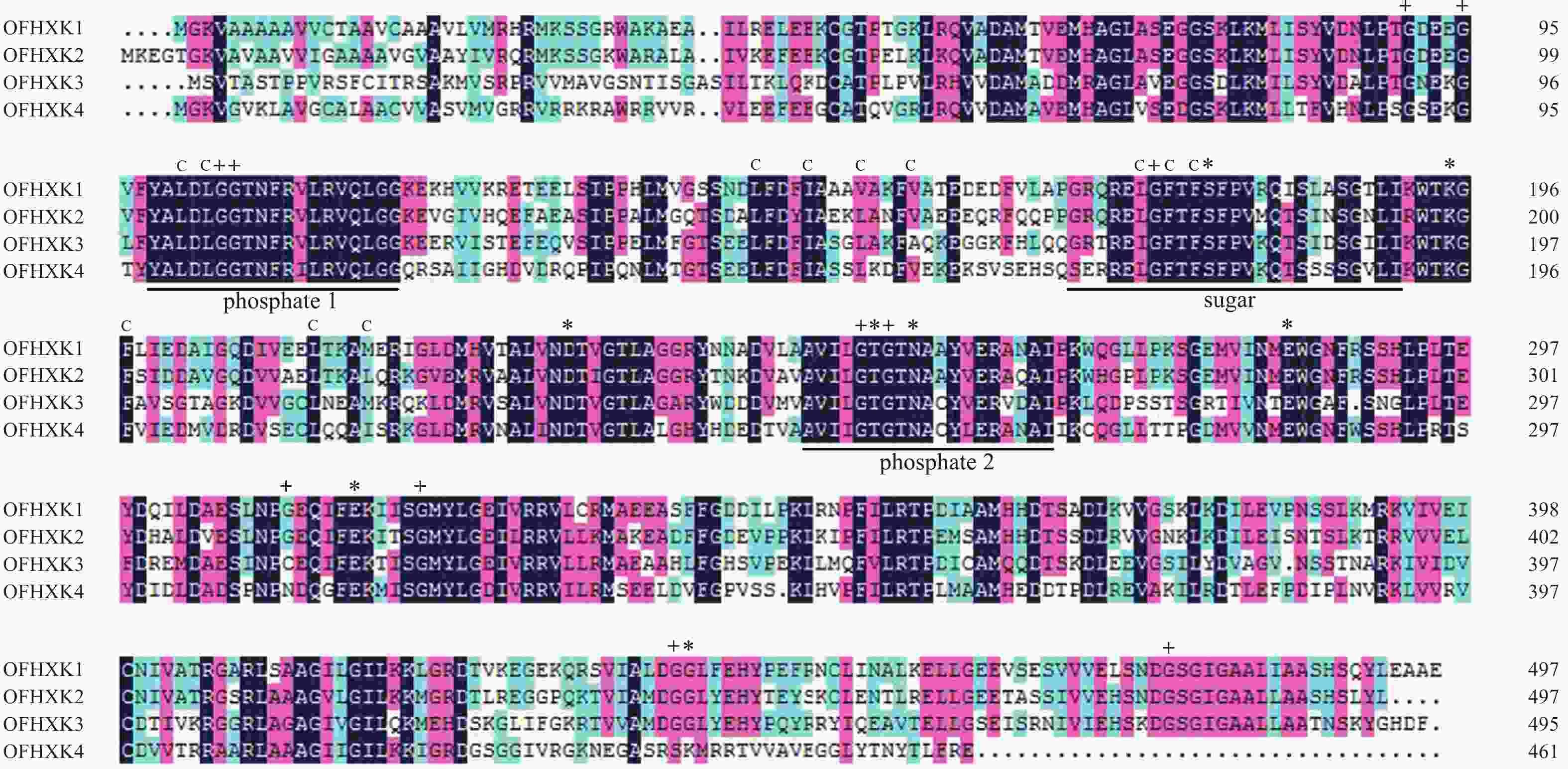
基因名称 相似性/% OfHXK1-YHG OfHXK1-YLL OfHXK1-JQG OfHXK2-YHG OfHXK2-YLL OfHXK2-JQG OfHXK3-YHG OfHXK3-YLL OfHXK3-JQG OfHXK4-YHG OfHXK4-YLL OfHXK4-JQG OfHXK1-YHG 100.00 OfHXK1-YLL 99.87 100.00 OfHXK1-JQG 99.87 100.00 100.00 OfHXK2-YHG 72.26 72.19 72.19 100.00 OfHXK2-YLL 72.21 72.14 72.14 94.42 100.00 OfHXK2-JQG 71.62 71.55 71.55 95.40 98.26 100.00 OfHXK3-YHG 58.38 58.45 58.45 59.90 58.37 58.03 100.00 OfHXK3-YLL 58.38 58.45 58.45 59.83 58.30 57.96 99.73 100.00 OfHXK3-JQG 58.38 58.45 58.58 59.62 58.23 57.83 99.19 99.33 100.00 OfHXK4-YHG 59.37 59.37 59.37 58.29 58.67 58.28 52.54 52.61 52.40 100.00 OfHXK4-YLL 59.63 59.63 59.63 58.86 58.66 58.44 52.65 52.73 52.50 99.71 100.00 OfHXK4-JQG 59.70 59.70 59.70 58.22 58.53 58.14 52.34 52.40 52.20 99.02 99.28 100.00 利用DNAMAN对‘堰虹桂’OfHXK1~OfHXK4氨基酸序列比对。由图1可知:不同OfHXKs间具有高度保守的结构域,均包含2个保守的磷酸化作用位点phosphate 1和phosphate 2以及1个底物结合位点sugar binding。预测OfHXK1与OfHXK2序列中均包含12个疏水通道和11个甘氨酸残基,而OfHXK3包含11个疏水通道和10个甘氨酸残基,OfHXK4则包含12个疏水通道和10个甘氨酸残基。与其他序列相比,OfHXK4氨基酸序列在其C端的腺苷结合位点处多出11个氨基酸残基(KNEGASRSKMR)。
-
利用ExPASy protparam预测分析OfHXKs家族蛋白质序列的基本理化性质,了解相对分子质量、氨基酸组成、等电点(PI)、不稳定系数、总平均亲水性等,结果见表3。以OfHXK1为例,该基因编码的氨基酸数目为498个,蛋白质相对分子量为53 881.09 Da,理论等电点为5.41,分子式为C2387H3857N653O719S21,氨基酸组成有20种,含有负电荷残基(Asp+Glu)66个,正电荷残基数(Arg+Lys)53个,总平均疏水性为0.055,不稳定系数为47.03,故推测OfHXK1为稳定蛋白质。结果显示:除OfHXK4为不稳定蛋白质外,其余均为稳定蛋白质。蛋白质信号肽预测使用SignalP在线软件默认条件下对OfHXKs氨基酸序列N-端开始前70位氨基酸进行信号肽预测分析,结果显示OfHXKs氨基酸序列均无信号肽存在。应用YLoc在线软件进行亚细胞定位预测,结果显示OfHXK1与OfHXK2均定位于细胞质中,而OfHXK3则定位于叶绿体,OfHXK4位于线粒体。
表 3 桂花OfHXKs蛋白质基本理化信息
Table 3. Information of OfHXK proteins in O. fragrans
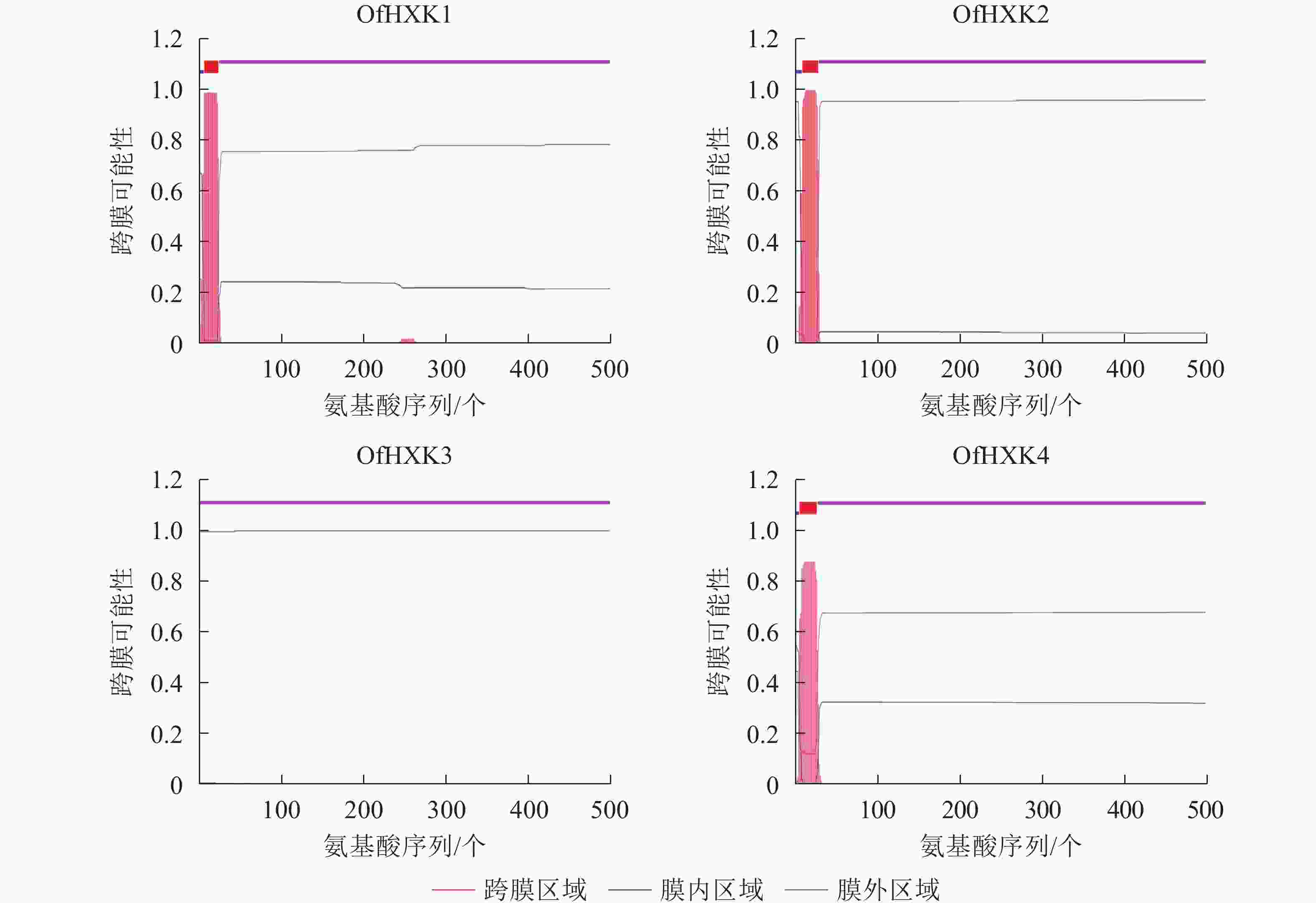
蛋白质 相对分子量 理论等电点 氨基酸数目/个 正电荷 负电荷 不稳定系数 总平均亲水指数 信号肽 亚细胞定位 OfHXK1 53881.09 5.41 498 53 66 37.03 0.055 无 细胞质 OfHXK2 54328.40 5.91 501 55 62 34.73 −0.077 无 细胞质 OfHXK3 53812.81 5.99 495 64 59 37.25 −0.023 无 叶绿体 OfHXK4 55894.38 6.58 510 61 63 46.37 −0.042 无 线粒体 使用TMHMM在线软件分析桂花OfHXKs蛋白质可能的跨膜区域,结果显示除OfHXK3外均具有跨膜区域(图2),OfHXK1、OfHXK2与OfHXK4分别在第7~24位氨基酸残基、第9~28位氨基酸残基和第5~24位氨基酸残基处具有跨膜区域,因此判断OfHXK1、OfHXK2与OfHXK4为跨膜蛋白质,而OfHXK3则不属于跨膜蛋白质。
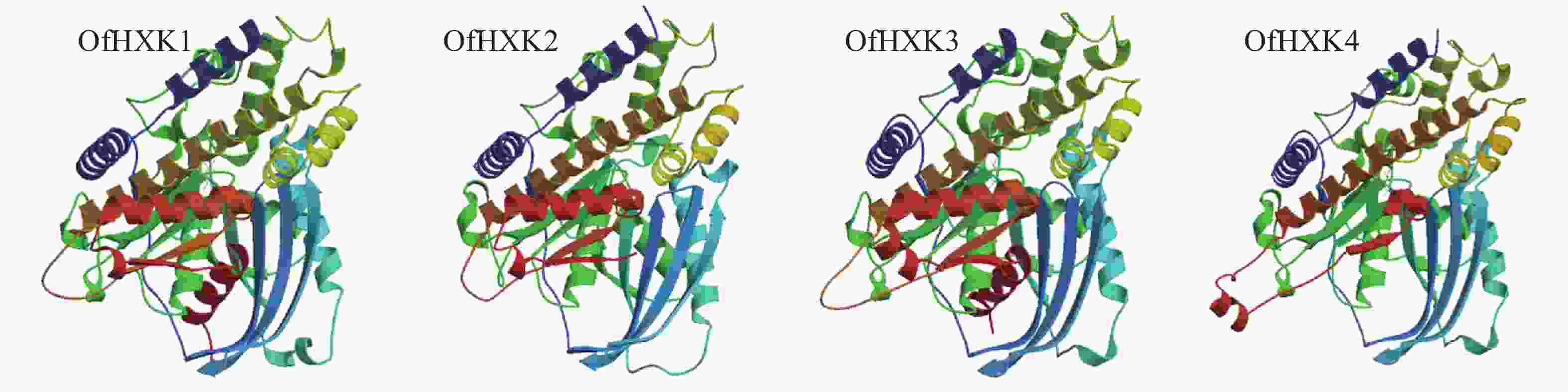
蛋白质的二级结构预测是为了判断目的基因所编码的氨基酸序列残基是处于α-螺旋、β折叠还是其他状态的二级结构。通过SOPMA在线预测分析,结果显示桂花OfHXKs蛋白质二级结构主要以α-螺旋和无规则卷曲为主(表4)。使用SWISS-MODEL进行三级结构预测,结果显示为图3。
表 4 桂花花瓣OfHXKs蛋白质二级结构预测
Table 4. Secondary structures of OfHXK proteins in O. fragrans
蛋白质 α-螺旋 β-转角 无规则卷曲 延伸链 氨基酸数目/个 占比/% 氨基酸数目/个 占比/% 氨基酸数目/个 占比/% 氨基酸数目/个 占比/% OfHXK1 239 47.99 31 6.22 162 32.53 66 13.25 OfHXK2 241 48.10 32 6.39 163 32.53 65 12.97 OfHXK3 208 42.02 31 6.26 188 37.98 68 13.74 OfHXK4 223 43.73 26 5.10 182 35.69 79 15.49 -
通过NCBI在线数据库比对,将4个桂花花瓣OfHXKs蛋白质序列分别进行同源性比对。Blastp比对结果显示:OfHXK1与油橄榄Olea europaea XP_022882495.1同源性为96.39%,与芝麻Sesamum indicum XP_022882495.1同源性为88.15%;OfHXK2与油橄榄XP_022843013.1同源性高达97.80%;OfHXK3与油橄榄XP_022884724.1同源性高达96.16%,而与阿月浑子Pistacia vera XP_031283181.1以及粉红钟花Handroanthus impetiginosus PIM98716.1同源性分别为82.70%与85.45%;OfHXK4与油橄榄XP_022873061.1同源性达到95%以上,说明OfHXKs具有高度保守性。
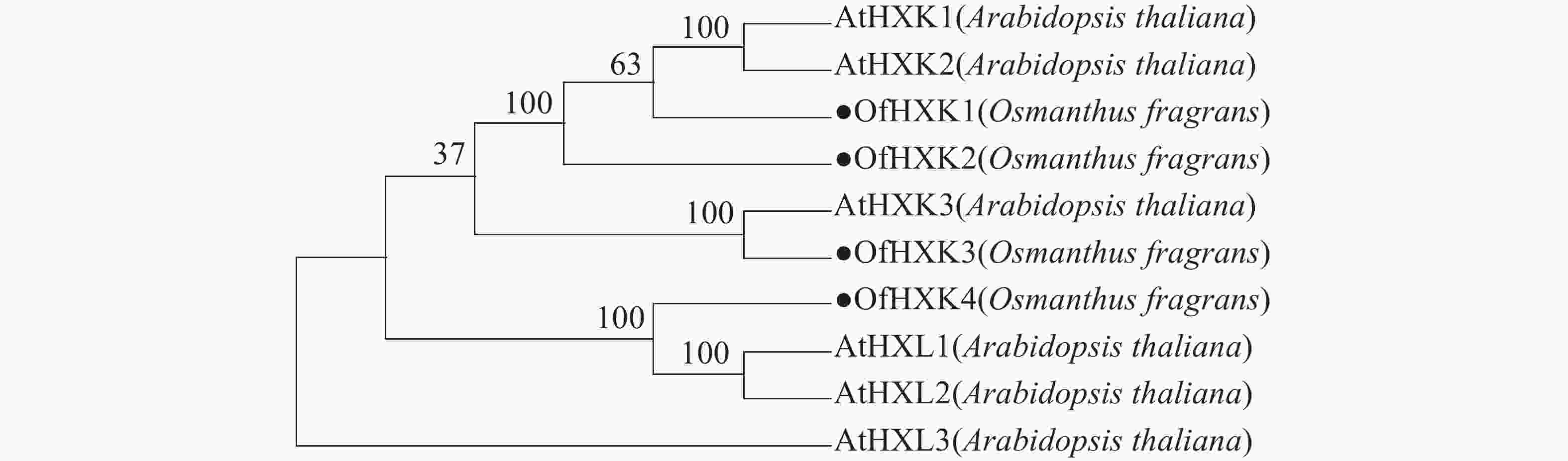
利用MEGAX软件对4个桂花花瓣OfHXKs蛋白质和从NCBI在线数据库中找到的拟南芥HXKs蛋白质序列进行多序列比对,以100% bootstrap支持度构建系统进化树进行分析,预测桂花OfHXKs的功能。由图4所示:所选择蛋白质序列分别为拟南芥AtHXK1(U28214)、AtHXK2(U28215)、AtHXK3(BT030472)、AtHKL1(NM_103929)、AtHKL2(NM_112895)和AtHKL3(NM_119945)。进化树分析结果表明桂花花瓣OfHXK1与OfHXK2关系较近,而OfHXK3和OfHXK4分别聚集在不同小支,由此推断OfHXKs蛋白质之间的功能可能具有一定区别。其中OfHXK1、OfHXK2与拟南芥AtHXK1、AtHXK2聚集在一支,而OfHXK3则与拟南芥AtHXK3关系较近,OfHXK4则与拟南芥AtHKL1、AtHKL2关系较近。
-
以桂花的顶壳期(S1)、铃梗期(S2)、初花期(S3)和盛花期(S4)花序以及1年生茎、2年生茎、嫩叶、成熟叶的cDNA为模板,以桂花ACT基因为内参,进行实时荧光定量PCR,检测不同品种桂花不同组织及不同发育阶段的OfHXKs基因表达情况。由图5可见:OfHXK1在‘堰虹桂’中的表达量呈现先上升后下降的趋势;在‘金球桂’中同样也在S3时期表达量最高,且显著高于‘堰虹桂’与‘玉玲珑’中相同时期的表达量(P<0.05);而OfHXK1基因在‘玉玲珑’中的表达呈现先上升后下降的趋势,于S2时期的表达量达到最高。OfHXK2基因在‘堰虹桂’中的表达量呈现逐步上升的趋势,于盛花期的表达量最高;在‘金球桂’中,OfHXK2的表达量随着开放过程未发生变化;OfHXK2的表达量在‘玉玲珑’中呈现先上升后下降的趋势。OfHXK3基因在3个品种的表达量随着花开放呈现先上升后下降的趋势,均在S2时期达到最高;S2时期‘金球桂’OfHXK3基因的表达量最高,‘玉玲珑’次之。OfHXK4基因在‘堰虹桂’和‘金球桂’中的表达量也是先上升后下降的趋势,在S2时期达到最高,而该基因在‘玉玲珑’中表达量逐渐下降。S2时期‘金球桂’OfHXK4基因的表达量最高,‘堰虹桂’次之。
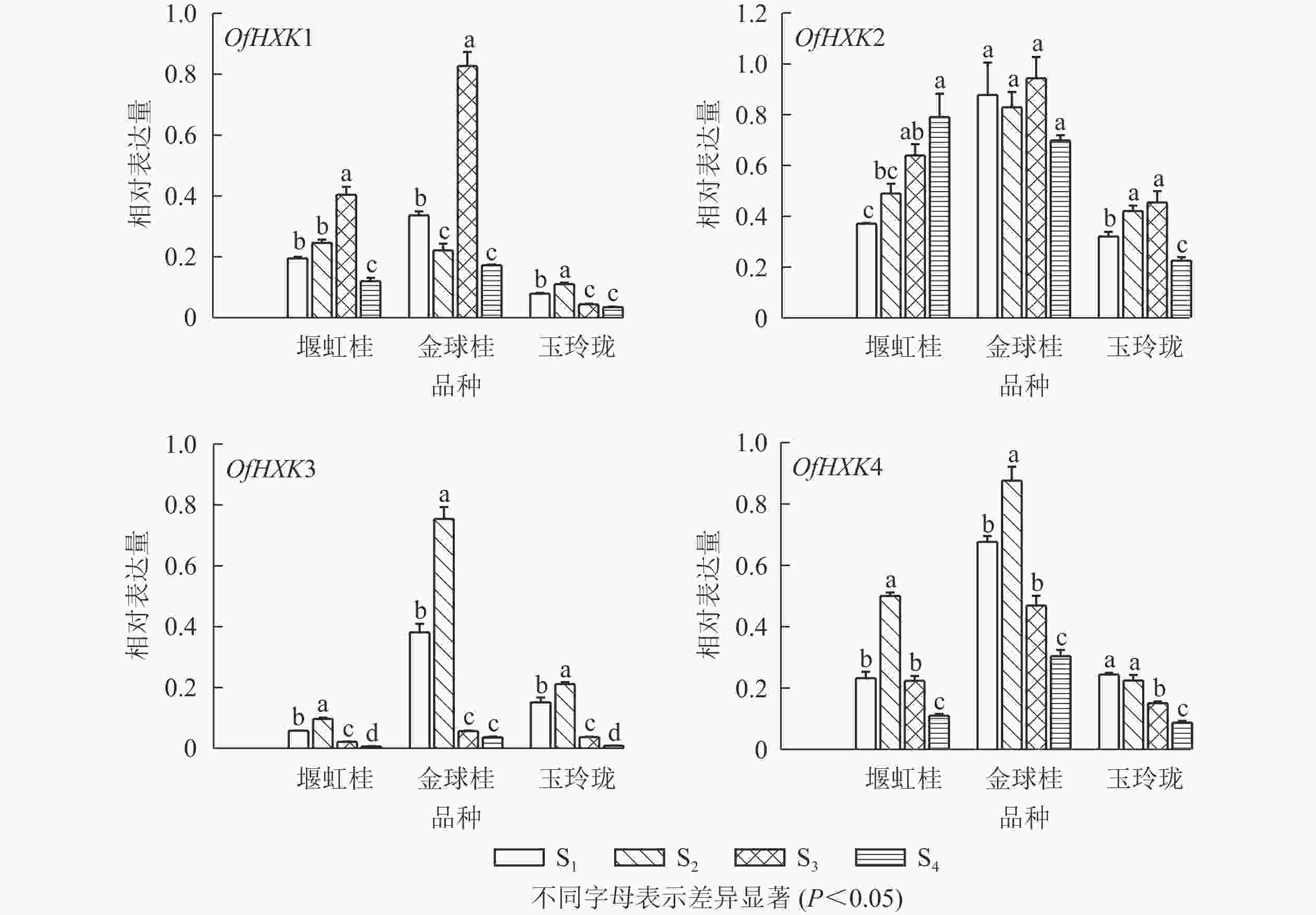
图 5 各品种桂花不同发育时期花序OfHXKs基因相对表达量变化
Figure 5. Expression changes of OfHXK genes during different inflorescence of different O. fragrans cultivars
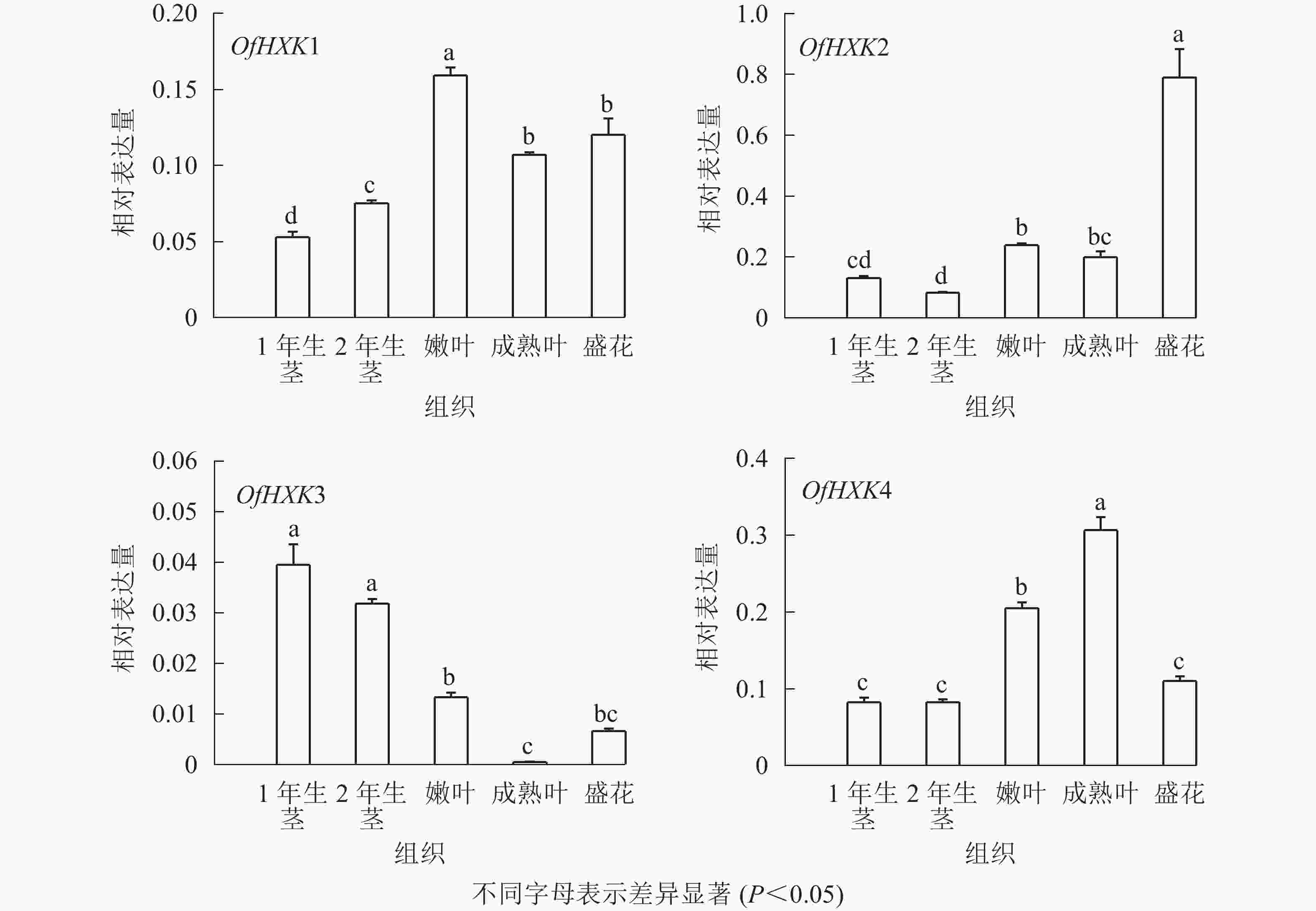
如图6所示:在不同组织中,OfHXK1在嫩叶中的表达量最高,在成熟叶和盛花期花序中表达量次之,在1年生茎中的表达量最低;OfHXK2在盛花期花序中的表达量最高,在2年生茎中的表达量最低;OfHXK3则在茎中表达量最高,在成熟叶中的表达量极低;OfHXK4在成熟叶中的表达量最高,嫩叶中表达量次之,显著高于1、2年生茎和盛开期花序中的表达量。
-
HXK参与糖代谢的同时也参与己糖信号的感受和传导[20-22],由于其重要的生物学功能,关于HXK蛋白的研究也逐渐成为热点[9, 14, 23-25]。本研究在桂花中发现HXK基因的4个同源基因,表明与其他植物一样[6],HXK基因也是以多基因家族形式存在桂花中,同一HXK基因家族成员序列在不同桂花品种中保守。桂花OfHXK1~OfHXK4基因推导的氨基酸序列均包含葡萄糖和ATP的结合位点,推测OfHXK1~OfHXK4均具有催化己糖磷酸化的功能。其中,OfHXK4氨基酸序列在其N端的腺苷结合位点处多出11个氨基酸残基,这与拟南芥中HKL1和HKL2腺苷结合位点处多10个和6个氨基酸残基的情况[10]类似。
进化树表明OfHXK1、OfHXK2与拟南芥AtHXK1、AtHXK2聚集在一支,已知AtHXK1和AtHXK2具有催化己糖磷酸化的作用[10],且具有糖感知和传导的功能[26],推测OfHXK1与OfHXK2同样具有催化己糖磷酸化和感知并转导糖信号的功能。OLSSON等[27]将植物HXK分为A类和B类,A类HXK具有质体信号肽,如AtHXK3;B类HXK具有N端膜锚定结构,如AtHXK1与AtHXK2。根据进化树推测OfHXK1和OfHXK2可能属于B类HXK,此外,核心的糖结合区域(LGFTFSFP-Q-L/I)在OfHXK1和OfHXK2序列中极其保守。OfHXK3与AtHXK3聚集于同一小支,拟南芥AtHXK3具有催化作用但无糖信号传导功能[28],推测无跨膜区域的OfHXK3属于A类HXK,具有催化作用但无糖信号传导功能。定位于线粒体中的拟南芥AtHKL1和AtHKL2属于酶催化活性不高的HXK亚型,其氨基酸序列C端的腺苷结合位点处多出若干个氨基酸残基[10],而推测聚在一支的OfHXK4有相同特点。
桂花OfHXK1~OfHXK4基因在1年生茎、2年生茎、嫩叶、成熟叶和盛开时花序中均有表达。其他物种的大部分HXK基因,如拟南芥AtHXK1~AtHXK3、AtHKL1和AtHKL2[10],番茄Lycopersicon esculentum LeHXK1~LeHXK4[11],水稻Oryza sativa OsHXK2~OsHXK9[8],枸杞Lycium barbarum LbHXK[29]和苹果Malus × domestica MdHXK1[30],在被检测的组织器官中均有表达。但是OfHXKs不同成员在不同组织中的表达水平呈现一定差异。OfHXK1和OfHXK2基因在叶和花中的表达量高于茎中,而OfHXK3基因在茎中的表达量最高,表明HXKs基因不同成员虽然磷酸己糖的功能存在冗余[10],但是不同成员在桂花不同组织中发挥的主导作用存在差异。依据进化树分析,OfHXK1与OfHXK2基因被推测具有糖信号感知和转导功能,可能参与调控桂花花色和花香相关次生代谢物质的生物合成,这可能是OfHXK1与OfHXK2基因在花中的表达量较高的原因之一。
本研究分析了3个桂花品种不同发育阶段花瓣中OfHXKs基因的表达情况,整体上‘金球桂’中OfHXKs基因表达量最高;随着花开放的进程,OfHXK1、OfHXK3和OfHXK4基因的表达量呈现先上升后下降的趋势。枸杞中研究发现,随着枸杞果实的发育,HXK基因表达量在色变期达到峰值,成熟期表达量又下降[29]。牡丹Paeonia suffruticosa花瓣中的研究发现:随着发育过程PsHXK1和PsHXK2基因在花瓣中的表达量呈现先增加后下降的规律,该变化规律与己糖含量有密切的关联[24]。植物组织器官中的糖含量在一定范围内增加可以促进HXK基因转录本和蛋白质的积累,植物体内己糖的磷酸化水平随之增加;超过一定阈值后,植物体内糖含量增加则会抑制HXK基因转录和蛋白质的积累[5]。这也就解释了大部分OfHXKs基因表达量先上升后下降的原因。桂花不同品种色素物质和香气挥发物含量不同[1-2],推测不同桂花品种对HXK己糖磷酸化和糖信号转导作用的需求不同,此外HXK不同成员还有功能冗余的特点,因此OfHXK2基因在3个桂花品种花瓣中的表达模式不同。
-
本研究根据桂花转录组Unigene序列分析,筛选得到4个HXK基因的同源基因。生物信息学分析结果表明4个基因分别编码461~510个氨基酸残基,同时推测OfHXK1~OfHXK4均具有催化己糖磷酸化的功能。进化树分析表明OfHXK1和OfHXK2与拟南芥AtHXK1与AtHXK2亲缘关系较近;OfHXK3与AtHXK3亲缘关系较近;OfHXK4与AtHKL1和AtHKL2亲缘关系较近。实时荧光定量PCR分析表明桂花OfHXK1~OfHXK4基因在1年生茎、2年生茎、嫩叶、成熟叶和花序中均有表达;OfHXK1、OfHXK3和OfHXK4基因随着花开放的进程,整体上表达量呈现先上升后下降的趋势,而OfHXK2基因在3个品种花序发育过程中表达模式不同。
Sequence and expression analysis of hexokinase gene family members in Osmanthus fragrans
-
摘要:
目的 研究桂花Osmanthus fragrans己糖激酶家族成员序列特征及表达变化规律。 方法 选取3个不同花色桂花品种‘堰虹桂’O. fragrans ‘Yanhong Gui’、‘玉玲珑’O. fragrans ‘Yulinglong’、‘金球桂’O. fragrans ‘Jinqiu Gui’,从转录组数据中筛选得到HXK同源基因,进行序列分析和系统进化树分析,并利用实时荧光定量PCR检测不同品种桂花不同组织及不同发育阶段的OfHXKs基因表达情况。 结果 筛选得到OfHXK1~OfHXK4基因,分析表明不同品种桂花OfHXK1、OfHXK3和OfHXK4基因核苷酸序列相似性均高于99%。OfHXKs基因编码461~510个氨基酸残基,均包含2个保守的磷酸化作用位点和1个糖结合位点。OfHXK1和OfHXK2具有N端膜锚定结构,与拟南芥Arabidopsis thaliana AtHXK1与AtHXK2聚为一支;无跨膜区域的OfHXK3与AtHXK3亲缘关系较近,推测二者具有催化作用但无糖信号传导功能;OfHXK4蛋白腺苷结合位点处多出11个氨基酸残基,与AtHKL1和AtHKL2亲缘关系较近。4个桂花HXK基因成员在桂花1年生茎、2年生茎、嫩叶、成熟叶和花序中均有表达。随着花开放的进程,整体上OfHXK1、OfHXK3和OfHXK4基因的表达量呈现先上升后下降的趋势,而OfHXK2基因在3个品种花序发育过程中表达模式不同。 结论 根据序列分析与进化树分析,推测桂花OfHXK1~OfHXK4均具有催化己糖磷酸化的功能,OfHXK1和OfHXK2具有糖信号感知和转导的功能。OfHXK1、OfHXK3和OfHXK4基因随花序发育呈现有规律升降的表达模式,可能与其参与糖类物质代谢有关。图6表4参30 Abstract:Objective This study aims to analyze the sequence characteristics and expression changes of hexokinase family members of Osmanthus fragrans. Method Based on the transcriptome data of O. fragrans cultivars ‘Yanhong Gui’, ‘Yulinglong’ and ‘Jinqiu Gui’, HXK homologous genes were selected for sequence analysis and phylogenetic tree analysis, and real-time fluorescence quantitative PCR was used to detect the expression of OfHXKs genes in different tissues and development stages of O. fragrans. Result Four HXK homologous genes OfHXK1−OfHXK4 were screened, and sequence identity of OfHXK1, OfHXK3 and OfHXK4 from different cultivars is more than 99%. OfHXKs encode 461−510 amino acid residues, including two conserved phosphorylation motifs phosphate 1 and phosphate 2, and one sugar binding motif. Sequence analysis and phylogenetic tree analysis showed that OfHXK1 and OfHXK2 have N-terminal membrane anchoring structures, which are grouped together with AtHXK1 and AtHXK2 from Arabidopsis thaliana. OfHXK3 without transmembrane region is closely related to AtHXK3, so it was speculated that OfHXK3 has catalytic effect but without sugar signaling function. OfHXK4 protein with 11 amino acid insertions at the adenosine binding domain, is closely related to AtHKL1 and AtHKL2. Four HXK gene members from O. fragrans are all expressed in the annual stem, biennial stem, tender leaves, mature leaves and inflorescence. With the development of flower, the expression levels of OfHXK1, OfHXK3 and OfHXK4 genes increase first and then decrease, while the expression patterns of OfHXK2 genes during flower development are different in the three cultivars. Conclusion According to sequence analysis and evolutionary tree analysis, it is speculated that OfHXK1−OfHXK4 all have the function of catalysing hexose phosphorylation, and OfHXK1 and OfHXK2 have the function of sugar sensing and signaling. The expression patterns of OfHXK1, OfHXK3, and OfHXK4 genes during the inflorescence development of the three varieties showed a regular increase and decrease with the process of flower opening, which may be associated with their functions in sugar metabolism. [Ch, 6 fig. 4 tab. 30 ref.] -
Key words:
- Osmanthus fragrans /
- hexokinase /
- gene cloning /
- expression analysis
-
表 1 OfHXKs基因表达分析所用引物序列
Table 1. Primer sequences of OfHXK genes of O. fragrans
基因名称 正向引物序列(5′→3′) 反向引物序列(5′→3′) OfHXK1 TTCTTCTTCGTCTGGCGTTCTG GTGCATTAACCCGCATATCCAGG OfHXK2 ACCTCCCTAAAAACAAGGAGAGTTG AGTATCCCGTCCCATTTTCTTTAGG OfHXK3 CACTTATTTGGTCACTCAGTTCCCG ACACACGTCTATGACAATCTTCCTC OfHXK4 GCACTCATTGCAGCCTCTCACT CTCACTCTGACAGTGACCGGCGTTA OfACT CCCAAGGCAAACAGAGAAAAAAT ACCCCACTACCAGAATCAAGAA 表 2 桂花OfHXKs核苷酸相似性比较表
Table 2. Comparison of nucleotide sequences of OfHXKs in O. fragrans
基因名称 相似性/% OfHXK1-YHG OfHXK1-YLL OfHXK1-JQG OfHXK2-YHG OfHXK2-YLL OfHXK2-JQG OfHXK3-YHG OfHXK3-YLL OfHXK3-JQG OfHXK4-YHG OfHXK4-YLL OfHXK4-JQG OfHXK1-YHG 100.00 OfHXK1-YLL 99.87 100.00 OfHXK1-JQG 99.87 100.00 100.00 OfHXK2-YHG 72.26 72.19 72.19 100.00 OfHXK2-YLL 72.21 72.14 72.14 94.42 100.00 OfHXK2-JQG 71.62 71.55 71.55 95.40 98.26 100.00 OfHXK3-YHG 58.38 58.45 58.45 59.90 58.37 58.03 100.00 OfHXK3-YLL 58.38 58.45 58.45 59.83 58.30 57.96 99.73 100.00 OfHXK3-JQG 58.38 58.45 58.58 59.62 58.23 57.83 99.19 99.33 100.00 OfHXK4-YHG 59.37 59.37 59.37 58.29 58.67 58.28 52.54 52.61 52.40 100.00 OfHXK4-YLL 59.63 59.63 59.63 58.86 58.66 58.44 52.65 52.73 52.50 99.71 100.00 OfHXK4-JQG 59.70 59.70 59.70 58.22 58.53 58.14 52.34 52.40 52.20 99.02 99.28 100.00 表 3 桂花OfHXKs蛋白质基本理化信息
Table 3. Information of OfHXK proteins in O. fragrans
蛋白质 相对分子量 理论等电点 氨基酸数目/个 正电荷 负电荷 不稳定系数 总平均亲水指数 信号肽 亚细胞定位 OfHXK1 53881.09 5.41 498 53 66 37.03 0.055 无 细胞质 OfHXK2 54328.40 5.91 501 55 62 34.73 −0.077 无 细胞质 OfHXK3 53812.81 5.99 495 64 59 37.25 −0.023 无 叶绿体 OfHXK4 55894.38 6.58 510 61 63 46.37 −0.042 无 线粒体 表 4 桂花花瓣OfHXKs蛋白质二级结构预测
Table 4. Secondary structures of OfHXK proteins in O. fragrans
蛋白质 α-螺旋 β-转角 无规则卷曲 延伸链 氨基酸数目/个 占比/% 氨基酸数目/个 占比/% 氨基酸数目/个 占比/% 氨基酸数目/个 占比/% OfHXK1 239 47.99 31 6.22 162 32.53 66 13.25 OfHXK2 241 48.10 32 6.39 163 32.53 65 12.97 OfHXK3 208 42.02 31 6.26 188 37.98 68 13.74 OfHXK4 223 43.73 26 5.10 182 35.69 79 15.49 -
[1] FU Jianxin, HOU Dan, WANG Yiguang, et al. Identification of floral aromatic volatile compounds in 29 cultivars from four groups of Osmanthus fragrans by gas chromatography-mass spectrometry [J]. Hortic Environ Biotechnol, 2019, 60(5): 611 − 623. [2] WANG Yiguang, ZHANG Chao, DONG Bin, et al. Carotenoid accumulation and its contribution to flower coloration of Osmanthus fragrans [J]. Front Plant Sci, 2018, 9: 1499. [3] GRANOT D. Role of tomato hexose kinases [J]. Funct Plant Biol, 2007, 34(6): 564 − 570. [4] 程建徽, 谢鸣, 蒋桂华, 等. 植物己糖激酶的信号转导作用[J]. 细胞生物学杂志, 2004, 26(6): 50 − 54. CHENG Jianhui, XIE Ming, JIANG Guihua, et al. The signaling role of hexokinase in plants [J]. Chin J Cell Biol, 2004, 26(6): 50 − 54. [5] WANG Xufeng, LI L M, YANG P P, et al. The role of hexokinases from grape berries (Vitis vinifera L.) in regulating the expression of cell wall invertase and sucrose synthase genes [J]. Plant Cell Rep, 2014, 33(2): 337 − 347. [6] KARVE R, LAURIA M, VIRNIG A, et al. Evolutionary lineages and functional diversification of plant hexokinases [J]. Mol Plant, 2010, 3(2): 334 − 346. [7] 张超, 王彦杰, 付建新, 等. 高等植物己糖激酶基因研究进展[J]. 生物技术通报, 2012, 28(4): 19 − 26. ZHANG Chao, WANG Yanjie, FU Jianxin, et al. Research advances in the hexokinase gene family in higher plant [J]. Biotechnol Bull, 2012, 28(4): 19 − 26. [8] CHO J, RYOO N, KO S, et al. Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.) [J]. Planta, 2006, 224(3): 598 − 611. [9] WANG Jingxue, WANG Xiaomin, GENG Siyu, et al. Correction to: genome-wide identification of hexokinase gene family in Brassica napus: structure, phylogenetic analysis, expression, and functional characterization [J]. Planta, 2018, 248(1): 171 − 182. [10] KARNE A, RAUH B L, XIA X, et al. Expression and evolutionary features of the hexokinase gene family in Arabidopsis [J]. Planta, 2008, 228(3): 411 − 425. [11] KANDELKFIR M, DAMARIWEISSLER H, GERMAN M A, et al. Two newly identified membrane-associated and plastidic tomato HXKs: characteristics, predicted structure and intracellular localization [J]. Planta, 2006, 224(6): 1341 − 1352. [12] JANG J, LEON P, ZHOU L, et al. Hexokinase as a sugar sensor in higher plants [J]. Plant Cell, 1997, 9(1): 5 − 19. [13] MOORE B D, ZHOU L, ROLLAND F, et al. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling [J]. Science, 2003, 300(5617): 332 − 336. [14] ZHOU L, JANG J, JONES T L, et al. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant [J]. Proc Nat Acad Sci USA, 1998, 95(17): 10294 − 10299. [15] PEGO J V, WEISBEEK P, SMEEKENS S, et al. Mannose inhibits arabidopsis germination via a hexokinase-mediated step [J]. Plant Physiol, 1999, 119(3): 1017 − 1023. [16] ZHENG Yanjun, TIAN Li, LIU Hongtao, et al. Sugars induce anthocyanin accumulation and flavanone 3-hydroxylase expression in grape berries [J]. Plant Growth Regul, 2009, 58(3): 251 − 260. [17] MIAO Huiying, WEI Jia, ZHAO Yanting, et al. Glucose signalling positively regulates aliphatic glucosinolate biosynthesis [J]. J Exp Bot, 2013, 64(4): 1097 − 1109. [18] 向其柏, 刘玉莲. 中国桂花品种图志[M]. 杭州: 浙江科学技术出版社, 2008. [19] ZHANG Chao, FU Jianxin, WANG Yiguang, et al. Identification of suitable reference genes for gene expression normalization in the quantitative real-time PCR analysis of sweet Osmanthus (Osmanthus fragrans Lour.) [J]. PLoS One, 2015, 10(8): 1 − 17. [20] 秦巧平, 张上隆, 徐昌杰. 己糖激酶与植物生长发育[J]. 植物生理学报, 2003, 39(1): 1 − 8. QIN Qiaoping, ZHANG Shanglong, XU Changjie. Hexokinase and development of plants [J]. Plant Physiol, 2003, 39(1): 1 − 8. [21] HALFORD N G, PURCELL P C, HARDIE D G, et al. Is hexokinase really a sugar sensor in plants [J]. Trends Plant Sci, 1999, 4(3): 117 − 120. [22] PERATA P, MATSUKURA C, VERNIERI P, et al. Sugar repression of a gibberellin-dependent signaling pathway in barley embryos [J]. Plant Cell, 1997, 9(12): 2197 − 2208. [23] GRANOT D, DAVIDSCHWARTZ R, KELLY G, et al. Hexose kinases and their role in sugar-sensing and plant development [J]. Front Plant Sci, 2013, 4: 44. [24] ZHANG Chao, ZHANG Lili, FU Jianxin, et al. Isolation and characterization of hexokinase genes PsHXK1 and PsHXK2 from tree peony (Paeonia suffruticosa Andrews) [J]. Mol Biol Rep, 2020, 47(1): 327 − 336. [25] HU Dagang, SUN Cuihui, ZHANG Quanyan, et al. Glucose sensor MdHXK1 phosphorylates and stabilizes MdbHLH3 to promote anthocyanin biosynthesis in apple [J]. PLoS Genet, 2016, 12(8): e1006273. doi: 10.1371/journal.pgen.1006273. [26] OLSSON T, THELANDER M, RONNE H. A novel type of chloroplast stromal hexokinase is the major glucose-phosphorylating enzyme in the moss Physcomitrella patens [J]. J Biol Chem, 2003, 278(45): 44439 − 44447. [27] MINET M, DUFOUR M, LACROUTE F, et al. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs [J]. Plant J, 1992, 2(3): 417 − 422. [28] 李浩霞, 杨斌, 尹跃, 等. 枸杞己糖激酶基因LbHXK的克隆及表达分析[J]. 西北植物学报, 2019, 39(6): 1009 − 1015. LI Haoxia, YANG Bin, YIN Yue, et al. Cloning and expression analysis of hexokinase gene (LbHXK) from wolfberry (Lycium barbarum Linn.) [J]. Acta Bot Boreali-Occident Sin, 2019, 39(6): 1009 − 1015. [29] 赵锦, 孙美红, 胡大刚, 等. 苹果己糖激酶基因MdHXK1的克隆与表达分析[J]. 园艺学报, 2015, 42(8): 1437 − 1447. ZHAO Jin, SUN Meihong, HU Dagang, et al. Molecular cloning and expression analysis of a hexokinase gene MdHXK1 in apple [J]. Acta Hortic Sin, 2015, 42(8): 1437 − 1447. [30] 张超. 葡萄糖调控牡丹切花花青素苷合成的分子机理[D]. 北京: 北京林业大学, 2014. ZHANG Chao. Molecular Mechanism of Glucose Regulating Anthocyanin Biosynthesis of Tree Peony Cut Flower[D]. Beijing: Beijing Forestry University, 2014. -

-
链接本文:
https://zlxb.zafu.edu.cn/article/doi/10.11833/j.issn.2095-0756.20200370






 下载:
下载: