-
绿色植物吸收光能主要依靠捕光色素蛋白复合体(light harvesting complex,LHC)完成,该复合体由LHC基因家族编码的蛋白结合叶绿素和类胡萝卜素组成,而这些LHC基因的表达是受光照调控的。强光可以抑制LHC基因的转录表达,同时能诱导一些本身具有光保护功能的LHC-Like基因表达,例如强光下OHPs(one helix proteins)、SEPs(stress enhanced proteins)和ELIPs(early light induced proteins)基因的表达增加[1-2]。ELIPs是核基因编码的受光诱导的叶绿体类囊体膜蛋白,最早发现于豌豆Pisum sativum黄化苗转绿过程。在此过程中ELIPs比其他的光诱导蛋白出现的更早,并在光合细胞器叶绿体发育完全后迅速消失[3-4]。ELIPs在蛋白结构上高度保守,目前已知的所有ELIPs都有3个α-螺旋跨膜结构,其中螺旋Ⅰ和螺旋Ⅲ与光系统中所有捕光叶绿素a/b结合蛋白相应部位有很高的序列同源性,因此ELIP蛋白被归入叶绿素a/b结合蛋白超家族,并根据其跨膜螺旋数量分为3个亚组[2,5]。目前,已经在拟南芥Arabidopsis thaliana[6]、葡萄Vitis vinifera[7]、紫花苜蓿Medicago sativa[8]、银杏Ginkgo biloba[9]等多种植物中克隆鉴定获得了ELIP基因。有关ELIP蛋白基因功能的研究也越来越得到研究者的重视。ELIPs是在有光的情况下产生的,在类囊体色素-蛋白质复合物的光保护或组装中起作用,也可在叶绿体到有色体的转换中发挥作用,对光合作用系统起到光修复作用[10-12]。一些ELIP基因在山墙藓Tortula ruralis中被干旱激活[13],在豌豆中能被紫外线B波段(UV-B)激活[14],在小麦Triticum aestivum中被低温激活[15]。推测ELIPs除了具有光保护功能外,可能具有适应与光抑制有关的非生物胁迫环境的功能[16]。此外,在拟南芥中发现,AtELIP1和AtELIP2基因的缺失会使其种子萌发率下降,且ELIP蛋白的抑制因子DAG1突变后,拟南芥种子萌发率则高于野生型,表明ELIP蛋白具有促进种子萌发的作用[17]。可见ELIPs在多种生理过程中可能具有重要作用。毛竹Phyllostachys edulis是重要的森林资源,生长迅速,材质优良,其光合作用一直倍受关注。人们已从竹子光合作用生理特性[18-19]、分子机理[20-25]等方面进行了大量研究。然而,对于毛竹的ELIPs尚缺乏研究。本研究以毛竹为研究对象,克隆ELIP基因,全面分析它的分子特征及表达模式,构建了PeELIP3表达载体并转化模式植物拟南芥,对基因功能进行了初步分析,以期为深入研究ELIP基因在竹子光保护中的生物学功能提供参考。
HTML
-
以实验室培养的毛竹实生苗为材料。培养条件:温度为18~25 ℃,光周期为16 h/8 h,光照强度为250~350 µmol·m−2·s−1,待长至6个月时进行光照处理。光强处理:将毛竹实生苗在黑暗适应24 h后分别移至于不同的光照强度(0、300、600、900、1 200和1 500 µmol·m−2·s−1)下,处理2 h后取样。强光处理:将毛竹实生苗置于1 200 µmol·m−2·s−1的强光下,分别在处理后0、0.5、1.0、2.0、4.0、8.0和12.0 h时取样。黄化苗处理:在黑暗条件下培养毛竹实生黄化苗,待长至1个月时,将毛竹黄化苗放置在250~350 µmol·m−2·s−1的光照下进行处理,分别在处理后0、0.5、1.0、4.0和8.0 h取样,并以室内正常光照(250~350 µmol·m−2·s−1)条件下生长的毛竹实生苗为对照(ck)。3次重复,每次重复的每个处理4~6株毛竹实生苗,所有处理均取苗顶端第3片叶为样本,液氮速冻后置于−80 ℃保存备用。
-
从前期毛竹基因组测序和RNA-Seq结果[22, 26]中筛选出与拟南芥ELIPs相似的基因,暂时将其认定为毛竹ELIP基因,并根据其序列设计特异性扩增引物(表1),进行目的基因扩增。利用Trizol法提取毛竹叶片的总RNA,并使用反转录试剂盒(Promage,美国)合成cDNA作为模板。PCR产物经琼脂糖凝胶DNA回收试剂盒完成回收,然后连接到pGEM-T easy Vector,转化大肠埃希菌Escherichia coli DH5α感受态细胞,筛选阳性克隆,在生工生物工程(上海)股份有限公司进行基因测序。
用途 基因名称 正向引物序列(5′→3′) 反向引物序列(5′→3′) 基因克隆 PeELIP1 ATGGCGACCAAGGTGGCCTT CTAGACGTTGACGAGCGGGGC PeELIP2 ATGGCGACGACCATGATGGC TTACACTACTAGTTTTAGACGTTGAC PeELIP3 ATGGCGACGACCATGATGAC TTAGATGTTGACGAACGGCGC 表达分析 PeELIP1 ATCATGTCCGCTGACGCCGA CTTTGTGCTAGACGTTGACGAGC PeELIP2 ACGACCATGATGGCCTCGAG TTGGGCGTCTCCGTTGGATC PeELIP3 GCGCATCTAGCCTGTGCAAT TTGTTCTGGGCCCTCACGAC Table 1. Sequence of primers
对克隆获得的基因及其编码的氨基酸序列进行分析,其中编码蛋白的基本理化性质通过ExPasy(http://www.expasy.org/)在线工具获取,相应功能结构域分析使用美国国家生物信息中心(NCBI)链接的在线数据库CD Search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi);利用Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant/#)和WoLF PSORT (https://www.genscript.com/psort/wolf_psort.html)预测亚细胞定位。此外,利用MEGA 7.0[27]提供的Clustal W工具对毛竹ELIP基因编码的氨基酸序列与其他物种ELIPs序列进行比对分析,且使用邻接法(neighbor-joining)构建系统进化树,重复次数1 000次,其他参数使用系统默认值。
-
根据获得的毛竹ELIP基因序列的非保守区域设计定量引物(表1)。利用qRT-PCR技术分析毛竹ELIP基因在不同光照处理条件下的表达模式,反应在qTower荧光定量PCR仪上进行,体系为10.0 μL:2 × SYBR Ⅱ Green 1 Master 5.0 μL;正/反向引物各0.3 μL (10 μmol·L−1);cDNA模板1.0 μL;H2O 3.4 μL。两步法进行PCR扩增:95 ℃ 6 min;95 ℃ 10 s,62 ℃ 10 s,40个循环。选择PeNTB为内参基因[28],基因的表达变化情况采用2–ΔΔCt法分析[29]。
-
通过PCR扩增得到两端分别带有BamHⅠ和XbaⅠ酶切位点的PeELIP3基因片段,酶切回收纯化后,将目的片段连接到pCAMBIA1301载体的多克隆位点,得到重组质粒pCAMBIA1301-PeELIP3。经测序验证正确后,将植物表达载体质粒利用电击法导入农杆菌Agrobacterium tumefaciens EHA105菌株,并经PCR验证正确后用于转化拟南芥。
-
利用农杆菌介导的浸花法[30]转化拟南芥,获得的T0代拟南芥种子经消毒后播种在含50 mg·L−1潮霉素的1/2 MS培养基中培养,初步挑选出具有抗性的转基因植株,并移栽到营养土中。提取T1代候选转基因植株的DNA,经PCR扩增PeELIP3基因序列,进一步鉴定转基因植株。在此基础上,筛选转基因植株至T3代,进行后续分析。同时,利用半定量RT-PCR检测PeELIP3在转基因拟南芥中的表达水平[23]。
-
参照韩志国等[31]方法,以3周的拟南芥植株为材料,利用IMAGING-PAM叶绿素荧光仪进行叶绿素荧光参数的测定,包括光系统Ⅱ最大光化学效率(Fv/Fm)、非光化学猝灭(NPQ)诱导曲线等。同时获取拟南芥的叶绿素荧光成像,分析野生型与转基因拟南芥植株之间的非光化学猝灭差异。
1.1. 材料与处理
1.2. 毛竹ELIP基因克隆与序列分析
1.3. 毛竹ELIPs基因的表达分析
1.4. PeELIP3植物表达载体构建
1.5. 转化拟南芥、筛选与鉴定
1.6. 转基因植株叶绿素荧光参数及荧光成像初步分析
-
经基因克隆、测序、序列分析和鉴定,从毛竹中获得3个ELIP同源基因,分别命名为PeELIP1、PeELIP2和PeELIP3。序列分析表明:3个PeELIPs基因的开放阅读框分别为498、540和549 bp,对应编码氨基酸大小分别为165、179和182个氨基酸,预测蛋白分子量为16.70、18.42和18.61 kDa。用DNAMAN分析3个PeELIPs基因编码的氨基酸序列,发现它们相互之间具有高度的相似性(80%以上),其中PeELIP2和PeELIP3之间达到91.1%。分别对3个PeELIPs蛋白的保守结构域进行预测,结果显示:它们的蛋白序列中都含有典型的捕光叶绿素a/b结合蛋白功能域,同时在该结构上都有3个α-螺旋跨膜结构,其中第1、3跨膜α-螺旋高度保守,且包括2个叶绿素结合位点:谷氨酸(Glu)残基和天冬酰胺(Asn)残基(图1),这一结构域是捕光叶绿素a/b结合蛋白的典型结构域[1, 5]。因此,3个PeELIPs均属于叶绿素a/b结合蛋白超家族成员。序列比对分析发现:3个PeELIPs与水稻Oryza sativa、玉米Zea mays等单子叶植物的ELIPs具有较高的相似性,同源性达72%以上,而与拟南芥等双子叶植物的ELIPs相似性较低,同源性低于67%。
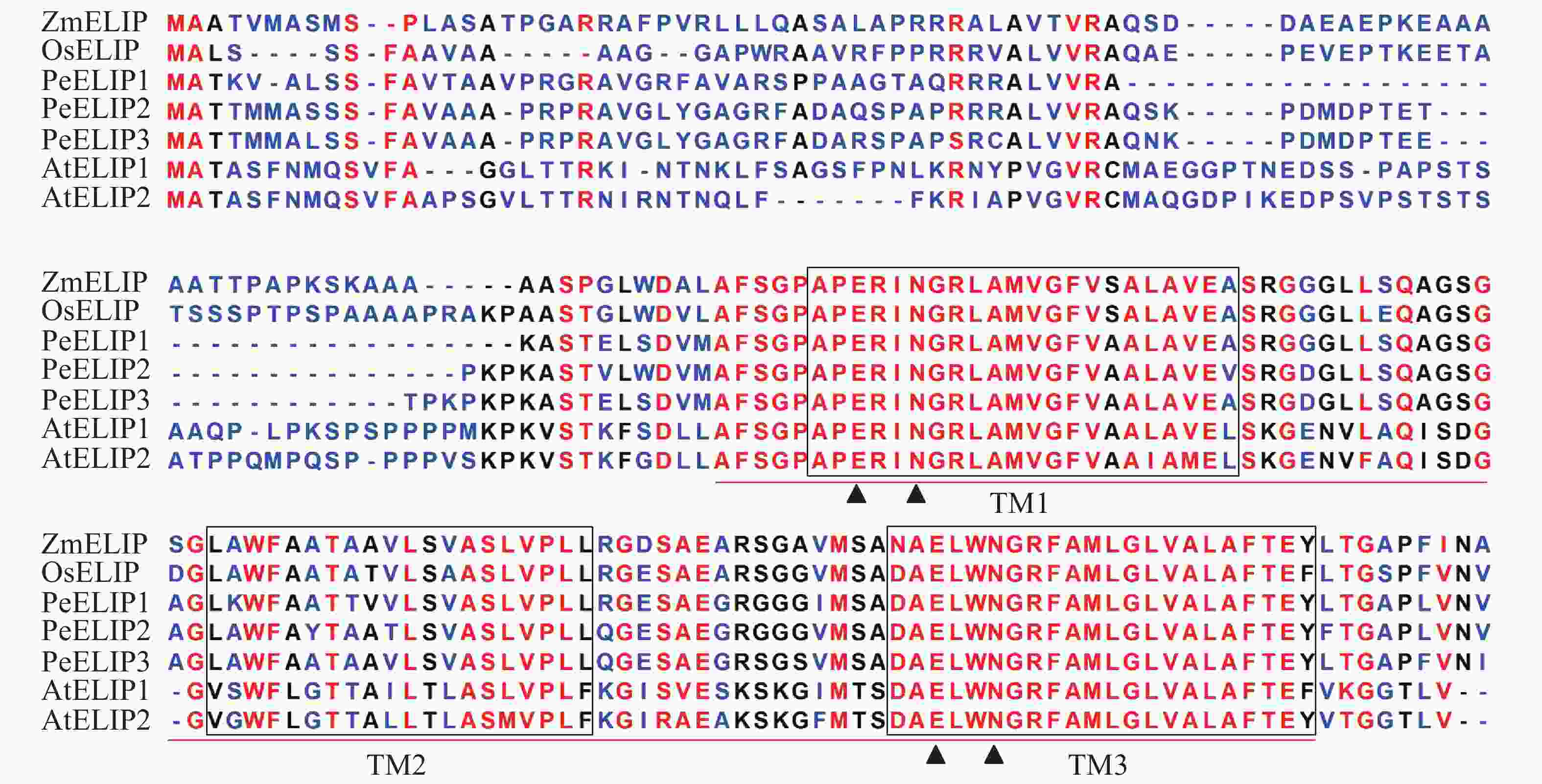
Figure 1. Alignment of the deduced amino acid sequences of ELIPs from Ph. edulis, A. thaliana, O. sativa and Z. mays
用Plant-PLoc在线软件进行亚细胞定位预测显示:PeELIP1和PeELIP2蛋白定位于叶绿体,而PeELIP3蛋白则定位在线粒体中。为了使预测结果更加准确,使用蛋白亚细胞定位预测专业软件WoLF PSORT在线对PeELIPs蛋白再次进行预测,结果显示:PeELIP1、PeELIP2和PeELIP3都最有可能定位于叶绿体中,进一步支持了它们属于叶绿素a/b结合蛋白超家族成员。
-
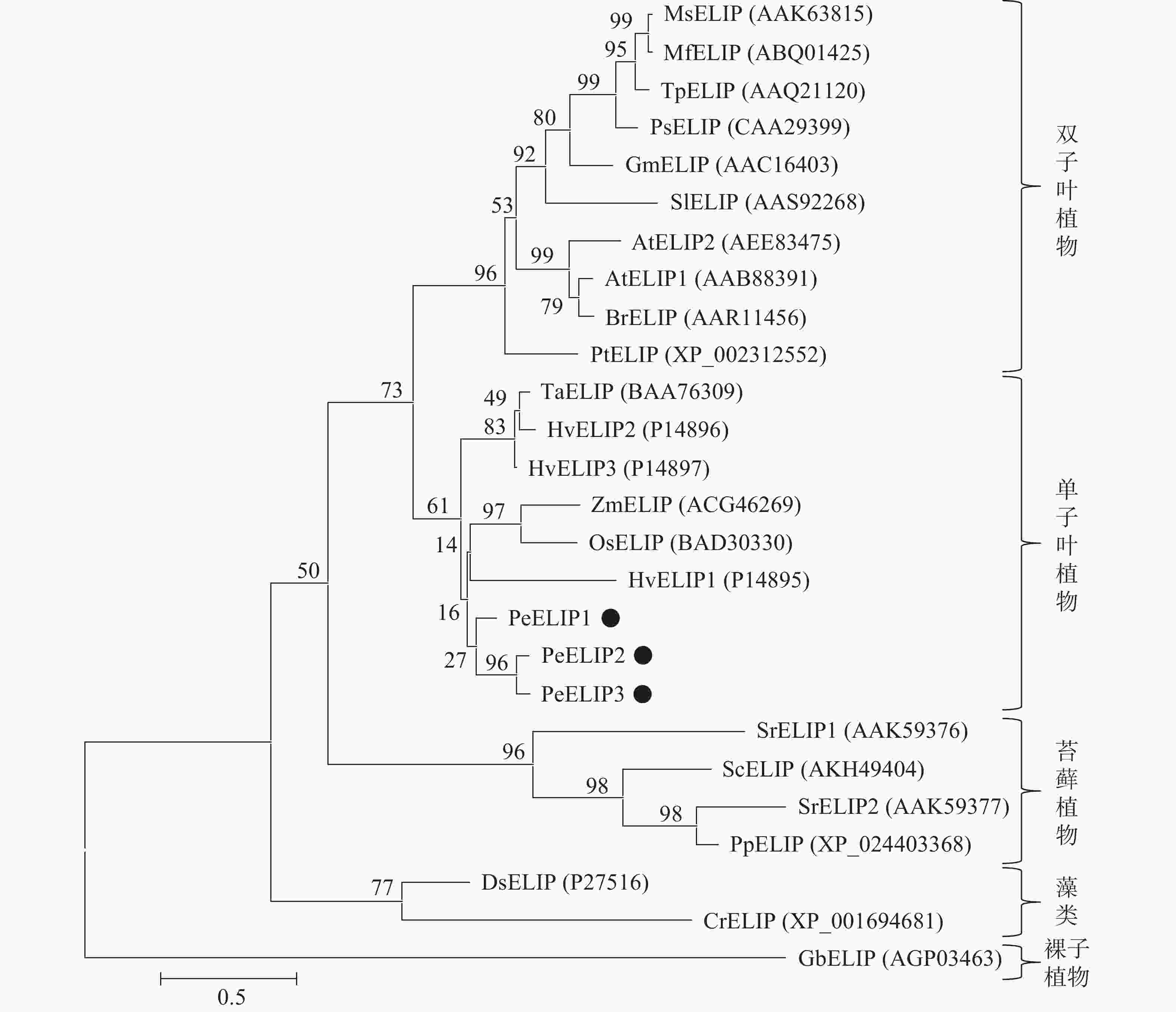
为揭示PeELIPs与其他物种ELIPs之间的进化关系,将PeELIPs与拟南芥、水稻、小立碗藓Physcomitrella patens等18个不同物种ELIPs的氨基酸序列进行比对分析,并构建进化树。结果(图2)表明:ELIPs进化可分为四大类,包括被子植物(双子叶植物和单子叶植物)、藻类、苔藓及裸子植物。毛竹ELIPs与水稻、玉米等单子叶植物ELIPs聚在一起,说明毛竹ELIPs与单子叶植物ELIPs蛋白具有较近的亲缘关系,然后是双子叶植物,与裸子植物、苔藓植物及藻类家族的亲缘关系较远,这与植物系统发育分类结果相一致。
-
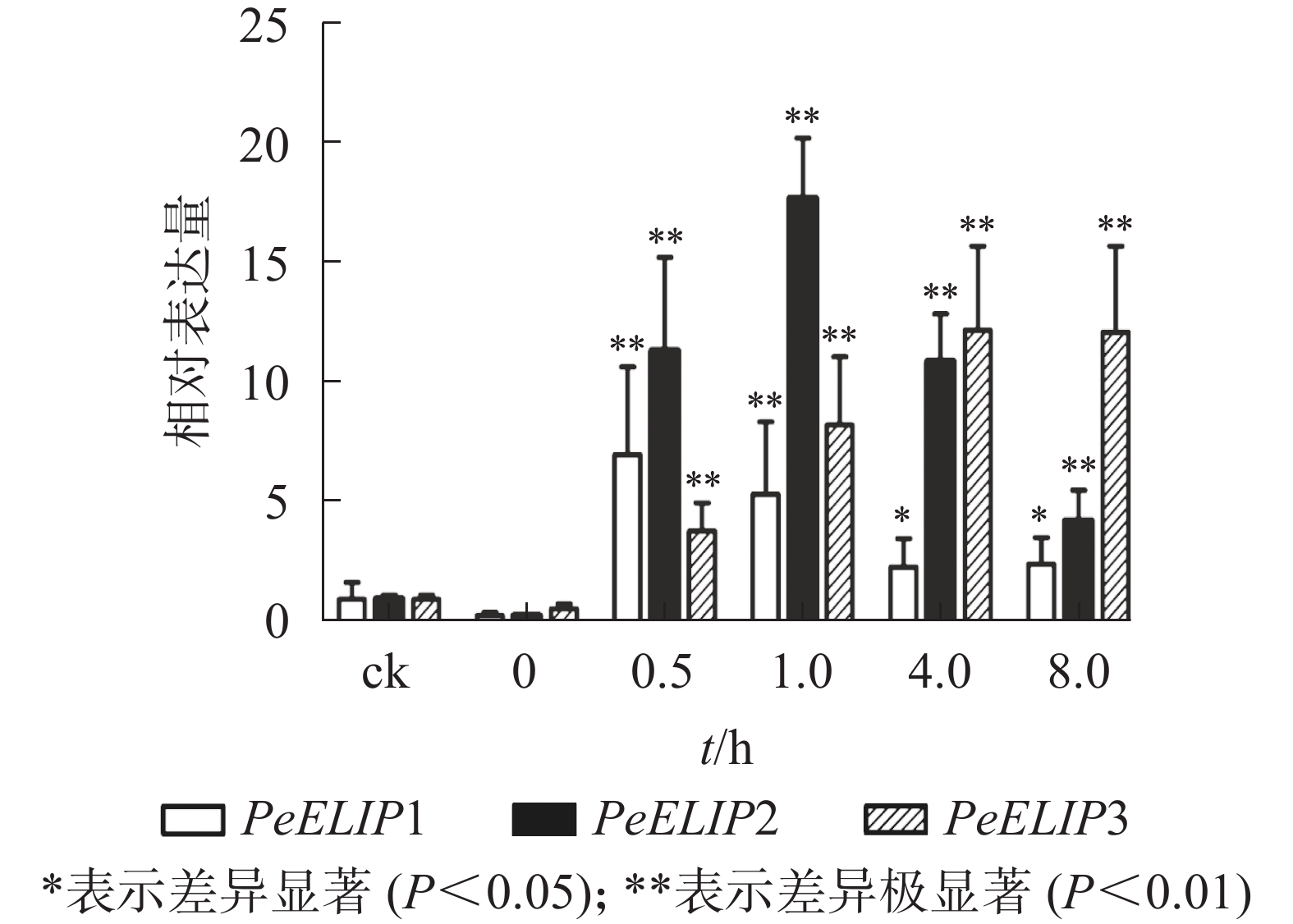
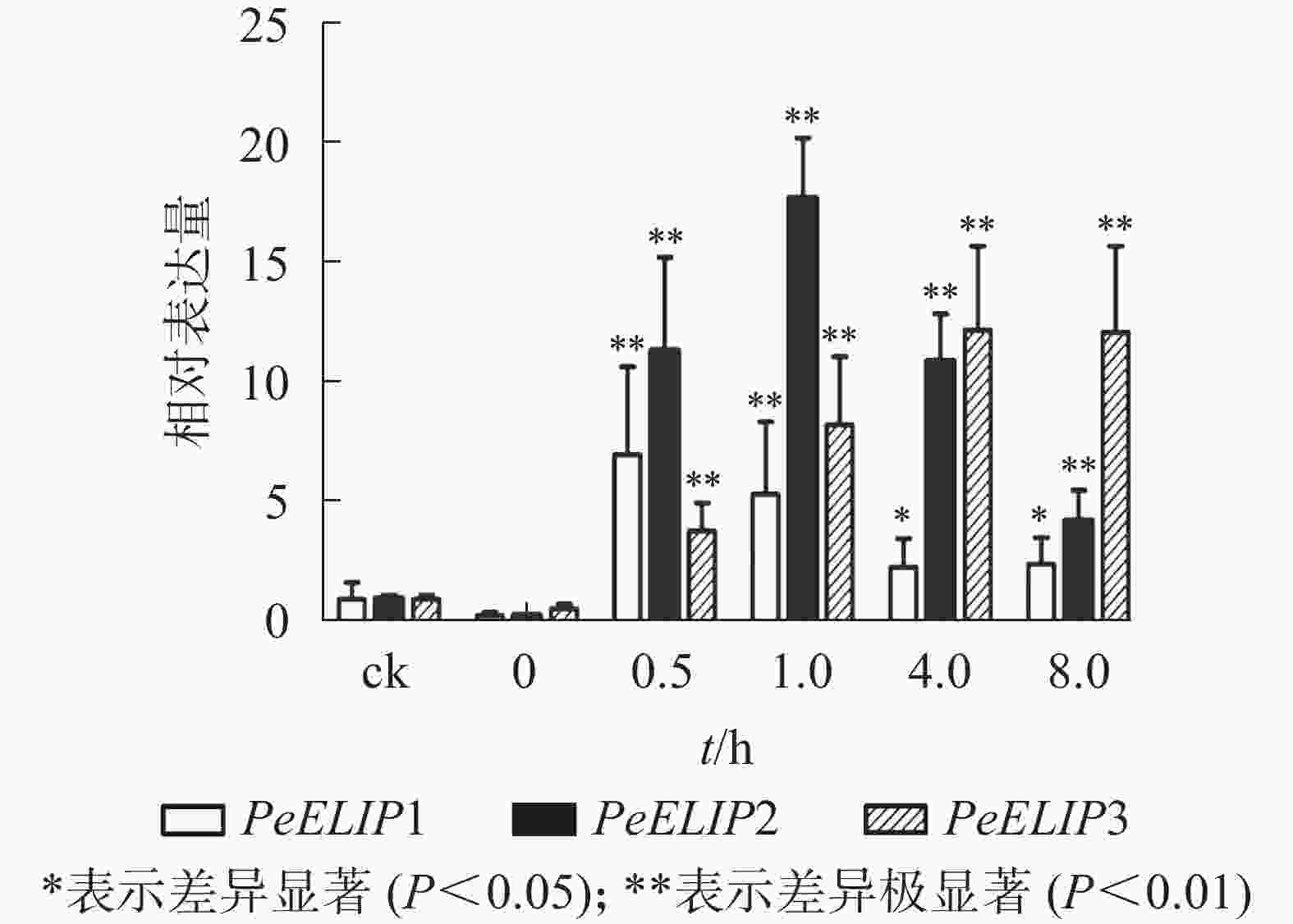
为分析光照对PeELIPs基因表达的影响,选择毛竹实生苗中的黄化苗为材料,进行光照处理(光照强度250~350 µmol·m−2·s−1),以绿色正常苗为对照(ck)。qRT-PCR结果(图3)显示:PeELIPs在毛竹黄化苗中仅检测到微量的表达,而在光照条件处理后黄化苗中3个PeELIPs的基因表达量均显著增加,其中PeELIP1和PeELIP2的表达量在光照处理的前1.0 h内快速增加,分别在0.5和1.0 h达峰值,与未光照处理的黄化苗相比,分别增加约21倍和58倍,随后出现下降趋势,至8.0 h时仍然高于ck,分别约为ck的2倍和4倍。而PeELIP3的表达量随光照处理时间的延长而上升,在4.0 h以后逐渐趋向平稳,在光照8.0 h后,其表达量约是对照的12倍,是黄化苗的20倍。
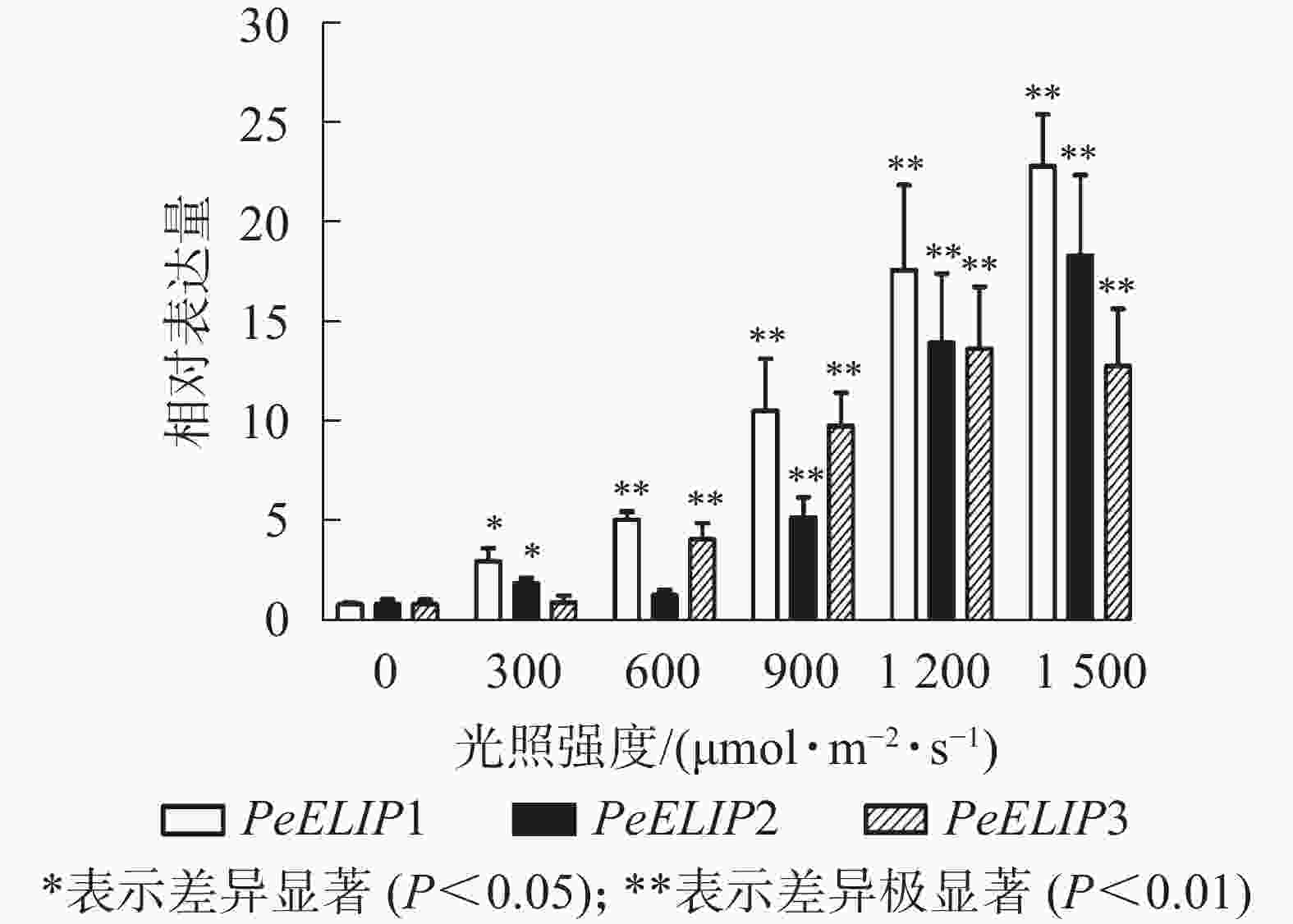
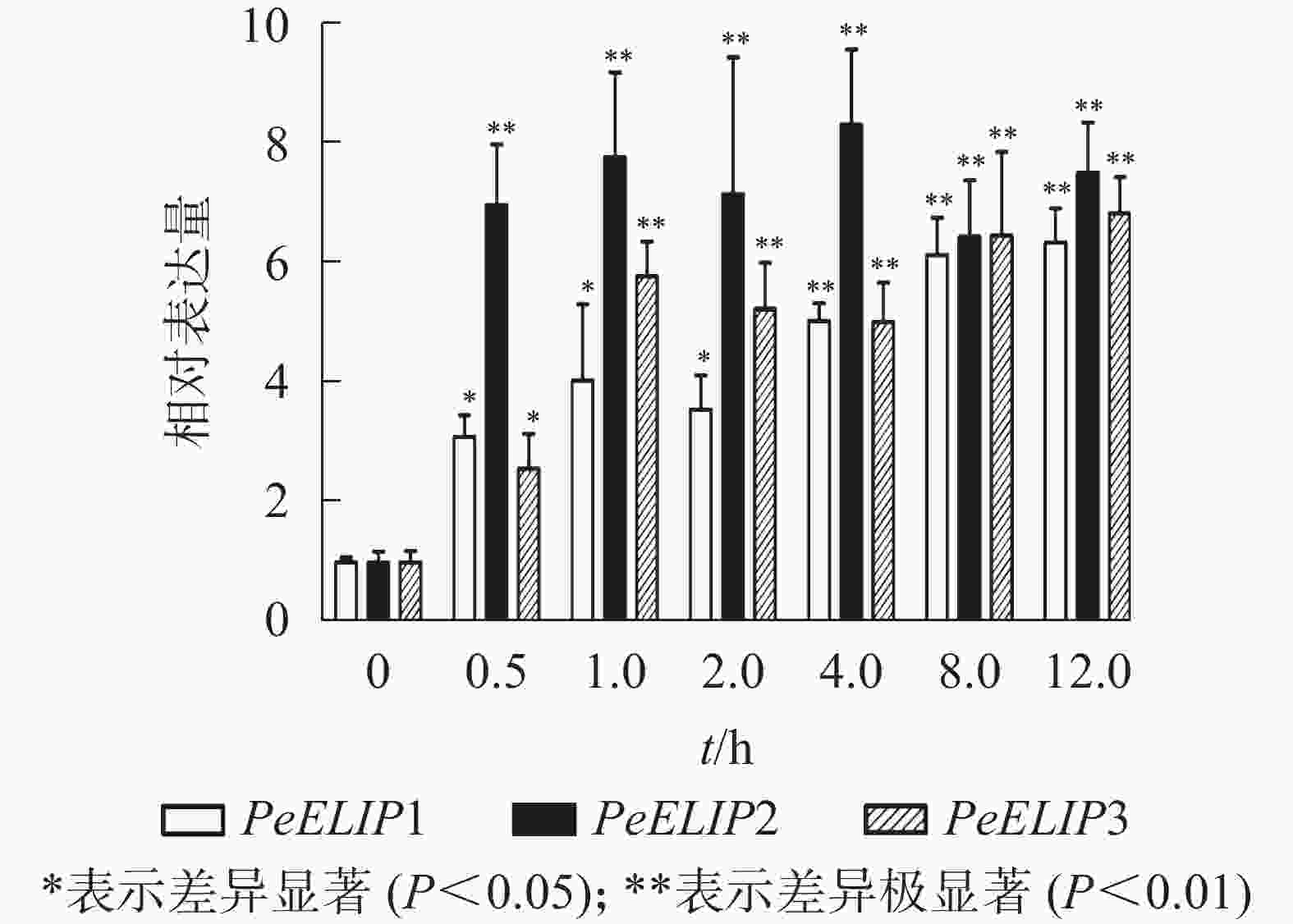
为进一步研究光照对PeELIPs基因表达的影响,选择不同光照强度(0、300、600、900、1 200和1 500 µmol·m−2·s−1)处理正常生长的毛竹实生苗叶片,检测其中PeELIPs的表达情况。qRT-PCR结果(图4)显示:随着光照强度增加,PeELIPs的表达量均比对照(0 h)增加,经1 500 µmol·m−2·s−1的光强处理2 h后,PeELIP1、PeELIP2和PeELIP3的表达量分别约是光照处理前的22、18和13倍,约是正常培养条件(300 µmol·m−2·s−1)下的7、9和12倍。在强光条件(1 200 µmol·m−2·s−1)下,随着处理时间的延长,PeELIPs的表达量都是先快速上升,随后变化不大,整体趋向平稳(图5)。毛竹黄化苗和正常苗中PeELIPs在不同光照强度、不同处理时间的表达变化表明,PeELIPs基因的表达受光照的诱导,且在光照初期的表达变化更为明显,进一步说明了PeELIPs编码的蛋白符合光早期诱导表达蛋白的特点。
-
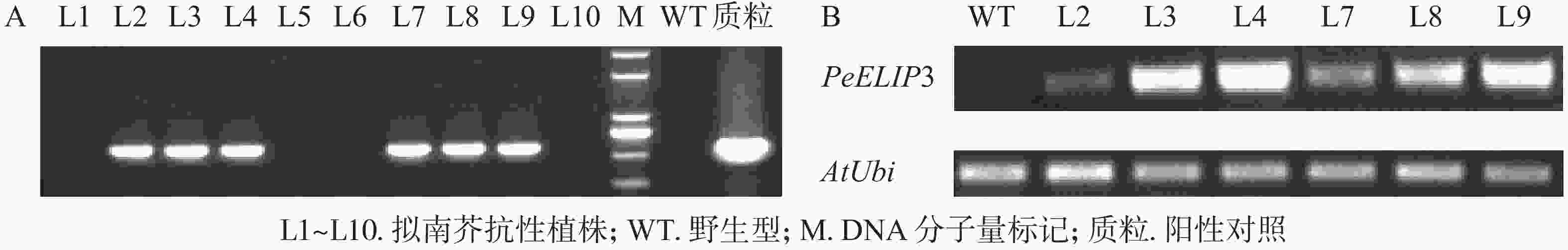
为验证PeELIPs基因的功能,选择其中1个基因PeELIP3构建了表达植物载体pCAMBIA1301-PeELIP3。将pCAMBIA1301-PeELIP3重组质粒通过农杆菌介导转化野生型拟南芥(Col-0),通过抗性筛选出的T1代拟南芥经基因组PCR和半定量RT-PCR鉴定,结果(图6A)显示:获得的10个抗性转基因拟南芥中6个株系(L2、L3、L4、L7、L8和L9)可检测到PeELIP3基因片段,而野生型拟南芥中没有检测到。随后对6个转基因株系继续进行培养,至T3代不分离后通过半定量RT-PCR进一步检测。结果(图6B)发现:PeELIP3基因在6个株系中均得到了表达,其中L3、L4、L8和L9株系中的表达量略高于L2和L7。这表明:PeELIP3基因已成功导入拟南芥中并得到转录表达,但不同株系中的表达量存在一定的差异。正常培养条件下,PeELIP3过量表达没有引起转基因拟南芥植株表型的变化。
-
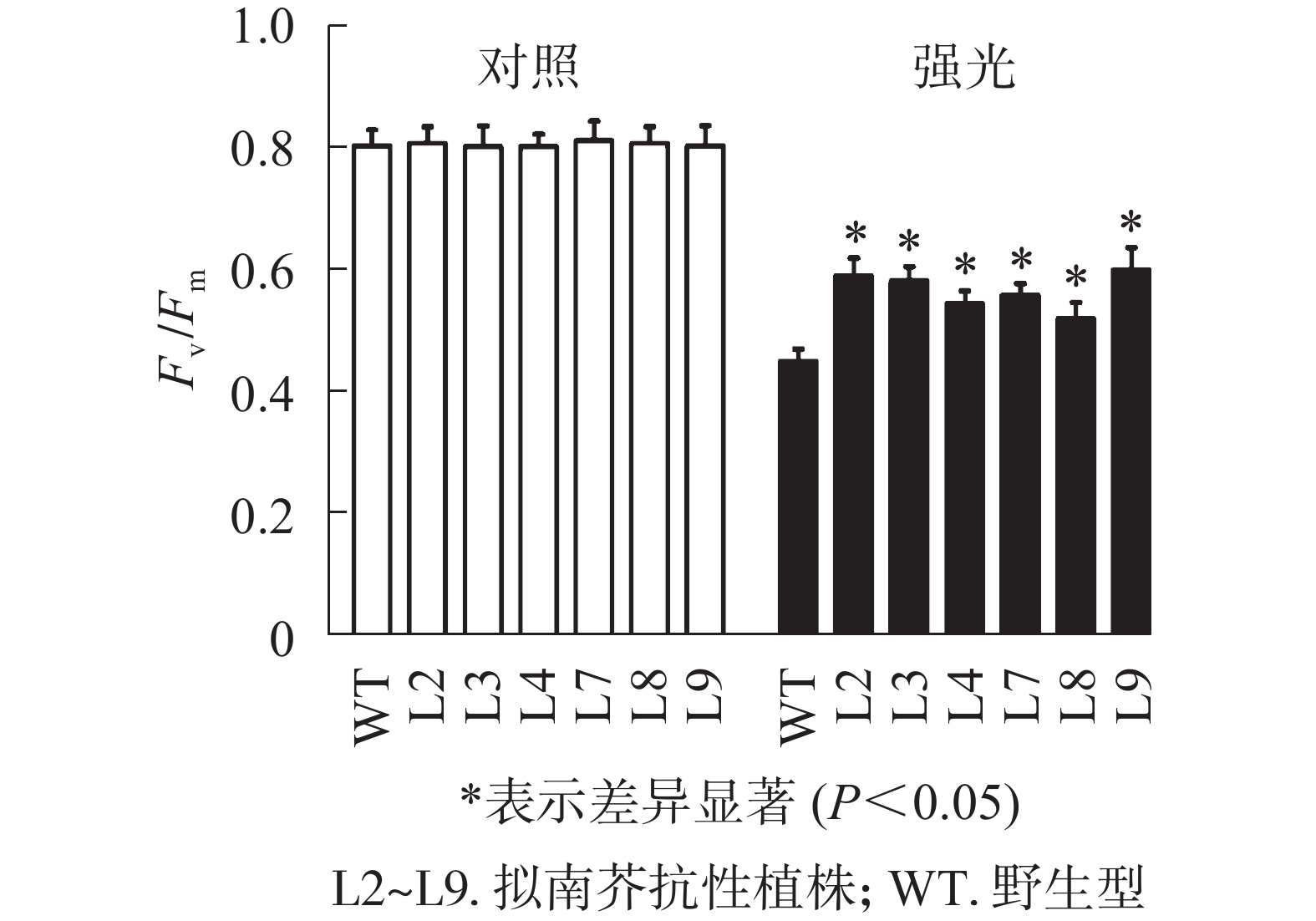
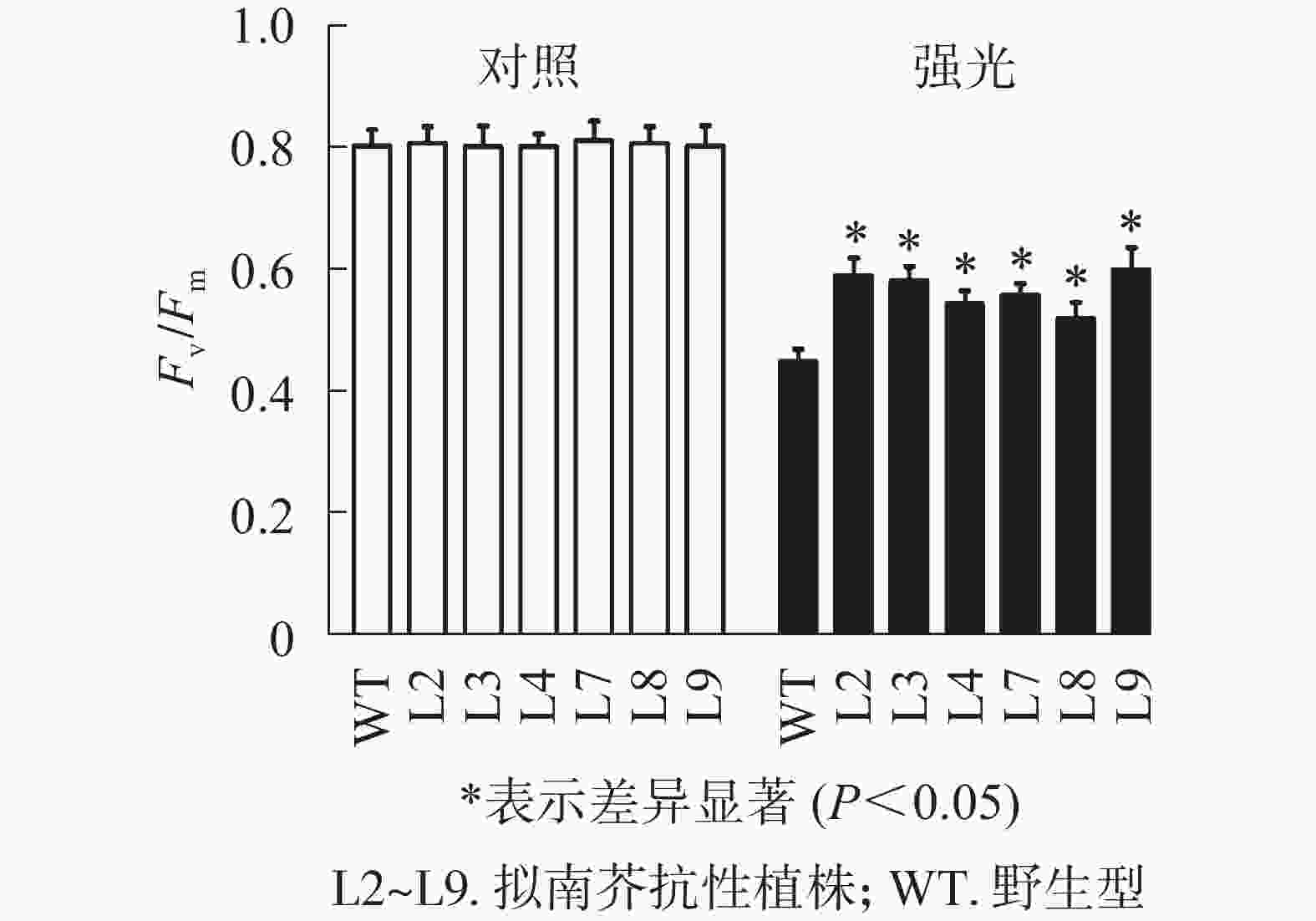
强光处理野生型和各转基因株系拟南芥,结果(图7)发现:经过4.0 h用1 200 µmol·m−2·s−1的强光处理后,野生型和转基因拟南芥的Fv/Fm都迅速下降,但两者的下降幅度存在显著差异,其中野生型拟南芥的Fv/Fm下降43.8%,而转基因拟南芥植株的下降幅度明显低于野生型拟南芥,仅下降25.1%~35.4%。由此表明:1 200 µmol·m−2·s−1强光处理后野生型和各转基因拟南芥均受到了胁迫,但过量表达PeELIP3可降低转基因拟南芥受强光胁迫的光抑制,以减缓Fv/Fm的下降程度。
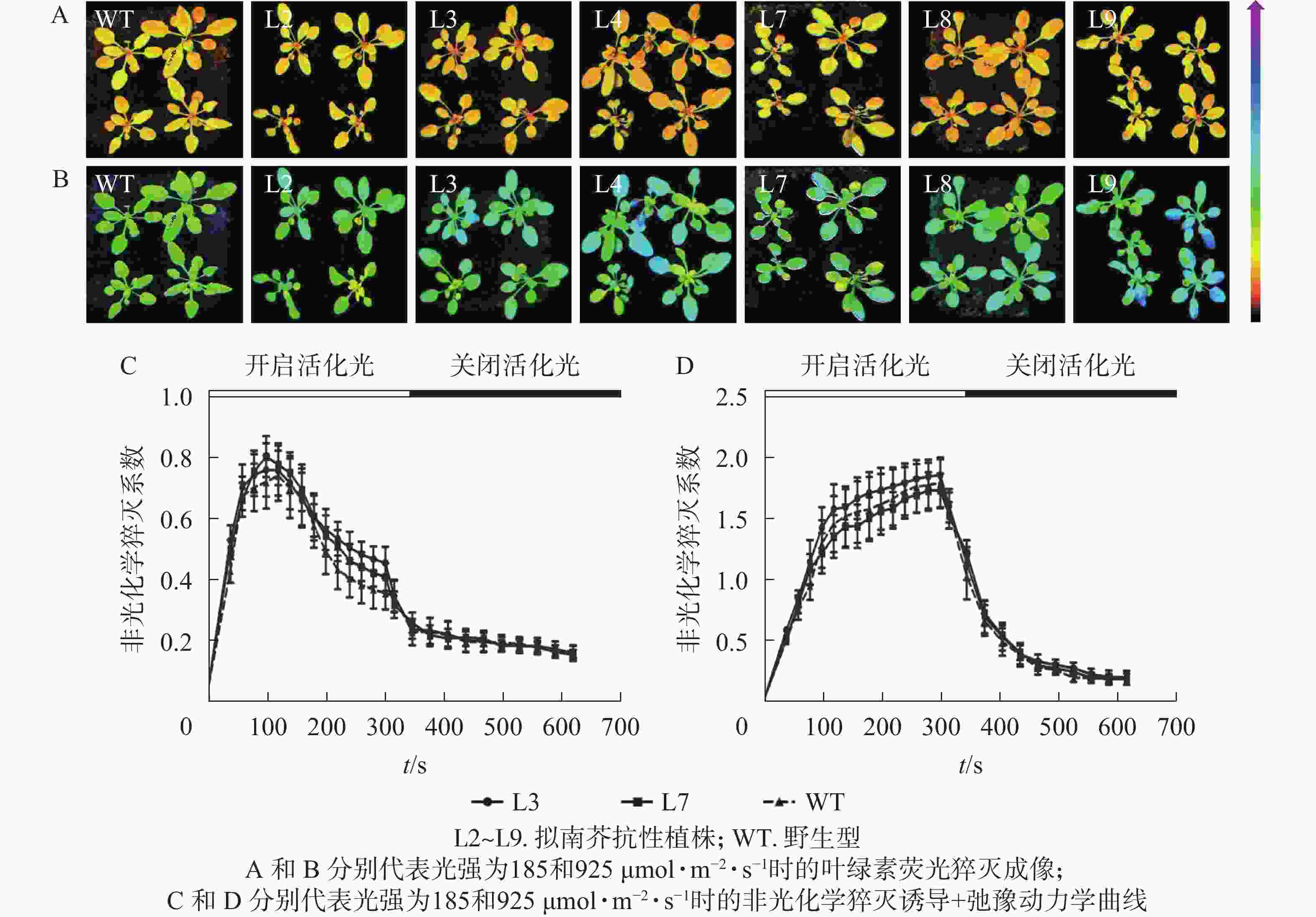
对PeELIP3转基因植株进行了非光化学猝灭成像和诱导+弛豫动力学分析。非光化学猝灭成像结果显示:在185 µmol·m−2·s−1的光强下,野生型和转基因拟南芥植株的非光化学猝灭成像均显示为黄色(图8A),当光强增加至925 µmol·m−2·s−1时,两者的非光化学猝灭成像均转变成青蓝色(图8B)。这说明在弱光或强光下,野生型和转基因拟南芥植株间的非光化学猝灭没有明显差异。同时,随机选取2个株系(L3和L7)进行非光化学猝灭诱导+弛豫动力学曲线分析。结果表明:在不同的活化光(弱光和强光)下,野生型与转基因拟南芥植株的非光化学猝灭诱导+弛豫动力学曲线没有明显差异,即185 µmol·m−2·s−1活化光下,野生型和转基因拟南芥的非光化学猝灭系数变化趋势都是先快速上升到达峰值,然后开始下降(图8C);当活化光光强设置为925 µmol·m−2·s−1时,两者的非光化学猝灭系数都是迅速上升,短时间内到达峰值,然后保持平稳(图8D)。在关闭活化光,进入暗弛豫后,两者的非光化学猝灭系数都在约5 min内快速弛豫(图8C和图8D)。以上结果表明:PeELIP3基因不影响转基因植株的非光化学猝灭,可能未参与热耗散相关的光保护途径。
2.1. PeELIPs基因克隆和序列分析
2.2. ELIPs系统进化分析
2.3. PeELIPs基因的表达分析
2.4. PeELIP3转基因拟南芥鉴定
2.5. PeELIP3转基因拟南芥叶绿素荧光参数分析
-
早期光诱导蛋白是核编码的叶绿体蛋白,在植物中广泛分布。但不同植物间编码ELIPs的基因数量存在较大差异,部分植物仅含有1个ELIP基因,如豌豆[14]和紫花苜蓿[8],而茶树Camellia sinensis[32]、番红花Crocus sativus[33]、杜鹃Rhododendron catawbiense[34]等部分植物中则有2~7个差异表达的ELIPs基因。本研究从毛竹中克隆得到了3个PeELIPs,它们分别编码的蛋白之间具有较高的相似性,均包含叶绿素a/b结合的结构域,属于捕光叶绿素a/b结合蛋白超家族成员。此外,PeELIPs蛋白与水稻、玉米等单子叶植物的ELIPs蛋白同源性较高,聚类在一个分支,但与苔藓植物和藻类的同源性相对偏低,这说明不同物种ELIPs蛋白存在较大差异。被子植物(双子叶和单子叶植物)、苔藓植物和藻类各自聚在一起,这表明ELIPs蛋白在相同进化等级的物种间相对比较保守,与物种的进化相一致。
大量研究结果表明:光照、温度、干旱等非生物胁迫影响ELIP基因表达调控。例如,17个旋蒴苣薹Boea hygrometrica ELIP基因中有7个基因受到干旱处理诱导[35],紫花苜蓿MsELIPs和杜鹃RcELIPs在低温胁迫中基因表达量增加[8, 34]。光照是诱导ELIP基因表达的主要因素。本研究中,毛竹黄化苗光照处理过程中,PeELIPs的表达量快速增加,在前1 h中达到顶峰,然后出现下降。这与番茄ELIP基因在其黄化苗转绿过程中的表达模式类似[36]。此外,3个毛竹PeELIPs的表达量都随着光照强度增加而上调,这表明光照诱导毛竹PeELIPs表达,其表达模式与拟南芥AtELIP1和AtELIP2[6]、葡萄VvELIP[7]、银杏GbELIP[9]基因类似。
研究[11]表明:拟南芥chaos突变体因为不能快速积累ELIPs蛋白,对强光和低温十分敏感,在强光胁迫下拟南芥chaos突变体植株叶片白化,表现出明显的光氧化现象,但将拟南芥ELIP基因在chaos突变体中组成性表达后,转基因拟南芥的强光耐受性又恢复到野生型状态,这表明ELIP蛋白具有光保护功能。盐藻Dunaliella salina DsCBR基因转化拟南芥elip突变体后,转基因拟南芥的强光耐受性提高到野生水平,这表明DsCBR基因与其高等植物ELIP基因同样具有光保护功能[37]。过量表达PeELIP3基因可减缓转基因拟南芥在强光胁迫下Fv/Fm的下降幅度,这表明PeELIP3可降低转基因拟南芥在强光胁迫中的光抑制程度,具有一定的光保护作用。另外,PeELIP1和PeELIP2的功能如何,有待深入研究。
热耗散过剩光能是植物重要的光保护途径,而叶绿素荧光参数非光化学猝灭系数能反映植物耗散过剩光能的能力,即光保护能力[38]。本研究中过表达PeELIP3基因不影响转基因拟南芥植株的非光化学猝灭,表明PeELIP3可能未参与非光化学猝灭的热耗散光保护途径。ELIPs光保护功能可能与其作为色素临时载体有关,强光胁迫下能加剧叶绿素结合蛋白的破坏,产生游离的叶绿素,而其容易被光敏化,形成叶绿素三聚体,叶绿素三聚体则与氧分子反应形成单线态氧,进而发生光氧化破坏,ELIP蛋白通过与叶绿素的短暂结合,防止游离叶绿素的积累,从而防止光氧化胁迫[11, 39]。同时,ELIP蛋白作为叶绿素传感器可调控叶绿素合成防止过多游离态叶绿素的积累[40]。因此,毛竹PeELIP3基因在光保护中的作用机制需要更多的研究去验证。









 DownLoad:
DownLoad: